 |
 |
 |
A specific domain for for the DNA2-NAM7 Helicase FamilyThe Dna2 yeast gene is required for DNA replication. This function was identified by screening for DNA replication mutants using a permeabilized cell replication assay (Budd and Campbell 1995). All the proteins retrieved by this profile have as a common feature: the interaction with nucleic acids. Several experiments have been performed with enzymes included in this group. The DNA2_YEAST enzyme, mutated at the position 1080, a lysin for a glutamate (Dna2K1080E), revealed that this residue is the ATP binding motif, because inactivates its ATPase and helicase activities and rendered mutant cells not viable. However, mutant cells expressing Dna2K1080T were capable of growth in media when lactate and glycerol were used as carbon source instead of glucose, suggesting that Dna2 appears to act in lagging strand metabolism on a role that is optimal with, but does not require, full helicase activity. Thus, the helicase activity of Dna2 may play a supportive role that becomes essential in rapidly growing cells (Bae, Kim et al. 2002). As in the DNA2_YEAST this domain is always associated to an ATP-Binding site, which is located between the domain and the N-termini of the protein, always closer to the domain.The domains and profiles importance is that the newly developed profile matches the DNA7/NAM7 protein family. Whereas DNA2 helicase is involved in DNA replication and NAM7 in mRNA turnover, they are both homologous to the transcription factor SMUBP-2, all of them retrieved by the profile. However, neither DNA2 nor NAM7 family members contain the DNA-binding domain of SMUBP-2, their direct role in transcription remains questionable (Kyrpides and Ouzounis 1999). Examples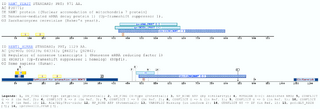
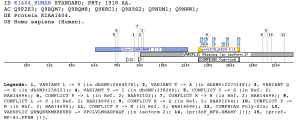

Taxonomic DistributionFungi: Saccharomyces cerevisiae, Schizosaccharomyces pombe, Yarrowia lipolytica. Plants: Arabidopsis thaliana, Oryza sativa. Vertebrates: Homo sapiens, Mus musculus. Invertebrates: Caenorhabditis elegans Caenorhabditis briggsae, Aquifex aeolicus. Bacteria: Mycoplasma pneumoniae Mycoplasma genitalium, Bacillus subtilis. Archaea: Methanococcus jannaschii, Pyrococcus abyssi. Viruses: Murine coronavirus, Human coronavirus. Insects: Drosophila melanogaster Anopheles gambiae Protista: Trypanosoma brucei, Plasmodium yoelii yoelii. PSI-BLAST against ensembl (Apis mellifera)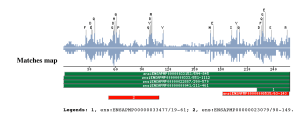
PSI-BLAST against ensembl (no taxonomic restriction)
Comparison against UNIPROT
Retrived sequences in UNIPROT (txt format) Downloadable filesProfileMultiple Alignment PSI-BLAST against ensembl (Apis mellifera) PSI-BLAST against ensembl (no taxonomic restriction) REFERENCESBae, S. H., D. W. Kim, et al. (2002). "Coupling of DNA helicase and endonuclease activities of yeast Dna2 facilitates Okazaki fragment processing." J Biol Chem 277(29): 26632-41. Budd, M. E. and J. L. Campbell (1995). "A yeast gene required for DNA replication encodes a protein with homology to DNA helicases." Proc Natl Acad Sci U S A 92(17): 7642-6. Kyrpides, N. C. and C. A. Ouzounis (1999). "Transcription in archaea." Proc Natl Acad Sci U S A 96(15): 8545-50. |