 |
 |
 |
Two new domains specific to the retrovirus- related Pol proteinsThe INTEGRASE_AD1 only matches retrovirus- related Pol proteins, mainly from plants, but also insects. Retroviral integrase is synthetised as a part of the POL polyprotein that contains an aspartyl protease, a reverse transcriptase, RNase H and integrase. POL polyprotein undergoes specific enzymatic cleavage to yield the mature proteins (Asante-Appiah and Skalka 1999). The retroviral integrase is the enzyme responsible for the insertion of a DNA copy of the viral genome into host DNA, an essential step in the replication cycle of viruses (Frankel and Young 1998). Integrases comprise three functional and structural domains: the central core domain, which contains the catalytic residues, an N-terminal zinc finger and a C-terminal DNA binding domain (Esposito and Craigie 1999).The integrase catalytic domain catalyzes a series of reactions to integrate the viral genome into a host chromosome. In the first step, it removes two 3' end nucleotides from each strand of the linear viral DNA, leaving overhanging CA-OH ends. In the second step, the processed 3' ends are covalently joined to the 5' ends of the target DNA. In the third step, which probably involves additional cellular enzymes, unpaired nucleotides at the viral 5' ends are removed and the ends are joined to the target site 3' ends, generating an integrated provirus flanked by five base-pair direct repeats of the target site DNA (van Gent, Elgersma et al. 1991). The domain INTEGRASE_AD1 is always located at the c-terminus of the domain Pfam: rve (PF00665), prf:INTEGRASE (PS50994) and the ipr: 4RNaseH_fold (IPR012337), which describe the integrase catalytic domain. In Q710T7_POPDE and, POLX_TOBAC, COPIA_DROME a short overlapping (approximately 15 residues) is present with the RVT_2 domain (PF07727). The domain INTEGRASE_AD2 is located at the C-terminus of the domain INTEGRASE_AD1. No annotations has been done in the matched region. Examples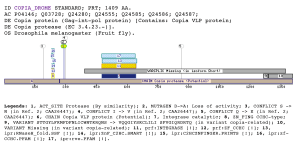

Taxonomic DistributionInsecta: Drosophila melanogaster, Aedes aegypti.Plants: Nicotiana tabacum (Common tobacco), Sorghum bicolor. Fungi: Saccharomyces cerevisiae, Pichia angusta, Cryptococcus neoformans. Vertebrata: Danio renio, Xenopus tropicalis.
REFERENCESAsante-Appiah, E. and A. M. Skalka (1999). "HIV-1 integrase: structural organization, conformational changes, and catalysis." Adv Virus Res 52: 351-69. Esposito, D. and R. Craigie (1999). "HIV integrase structure and function." Adv Virus Res 52: 319-33. Frankel, A. D. and J. A. Young (1998). "HIV-1: fifteen proteins and an RNA." Annu Rev Biochem 67: 1-25. van Gent, D. C., Y. Elgersma, et al. (1991). "DNA binding properties of the integrase proteins of human immunodeficiency viruses types 1 and 2." Nucleic Acids Res 19(14): 3821-7. |