 |
 |
 |
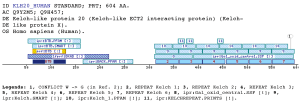
BKAD: BTB & Kelch associated domain (BKAD)The kelch motif is an ancient and evolutionarily-widespread sequence motif of 44-56 amino acids in length. It occurs as five to seven repeats that form a -propeller structure (Prag and Adams 2003). This module appears in many different polypeptide contexts and contains multiple potential protein-protein contact sites. Five subgroups of kelch-repeat proteins have been defined accordingly to the position of the kelch -propeller and the presence of other protein domains within the primary sequences (Adams, Kelso et al. 2000). One of this subgroups, is defined as N-dimer, C-propeller proteins, which contain an N-terminal BTB/POZ (Broad-Complex, Tramtrack and Bric a brac) domain and four to six kelch motifs located within the C-terminal region.The BTB domain (IPR000210) is present near the N-terminus of a fraction of zinc finger (zf-C2H2) proteins and in proteins that contain the Kelch motif such as Kelch and a family of poxvirus proteins, as well as in several proteins with diverse functions. The BTB/POZ domain mediates homomeric dimerisation and in some instances heteromeric dimerisation. This wide distribution is what do likely that the BTB domain has a wide distribution in size (Deweindt, Albagli et al. 1995) Previous studies have allowed determining the interaction between some of the proteins included in the group matched by this domain, with Topoisomerase I (TOP1). BTBD1 and BTBD2 were mapped by deletion analysis to identify amino acid sequences required for interaction with TOP1. The N-terminal deletions of ~36 % of BTBD1 (177 residues) or 43% BTBD2 (228 residues) do not eliminate binding to TOP1. In contrast, C-terminal deletions of 112 and 33 residues of BTBD1 (Q9H0C5) and BTBD2 (Q9BX70), respectively, eliminated binding to TOP1 in the both cases. The domain here proposed is within the region that is related to the interaction, so it could be involved. Nevertheless BTBD1 and BTBD2 and their interaction with TOP1 are underway (Xu, He et al. 2004). The domain BKAD is always found associated with the BTB domain and the Kelch repeat and is located betwixt them. A large number and type of proteins contain this arrangement of motifs, but if the Kelch motifs are not present in a BTB protein, the BKAD is not present either. Examples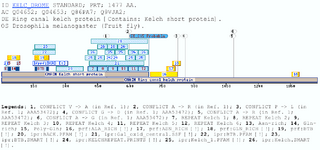


Taxonomic DistributionVertebrates: Homo sapiens, Mus musculus, Rattus norvegicus. Insecta: Anopheles stephensi, Drosophila melanogaster. Invertebrates: Caenorhabditis elegans, Plants: Arabidopsis thaliana, Oryza sativa. Protozoa: Cryptosporidium hominis. Viruses: Vaccinia virus, Variola virus. PSI-BLAST against ensembl (Apis mellifera)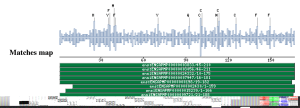
PSI-BLAST against ensembl (no taxonomic restriction)
Comparison against UNIPROT
Retrived sequences in UNIPROT (txt format) Downloadable filesProfileMultiple Alignment PSI-BLAST against ensembl (Apis mellifera) PSI-BLAST against ensembl (no taxonomic restriction) REFERENCESAdams, J., R. Kelso, et al. (2000). "The kelch repeat superfamily of proteins: propellers of cell function." Trends Cell Biol 10(1): 17-24. Deweindt, C., O. Albagli, et al. (1995). "The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: a novel function for the BTB/POZ domain as an autonomous repressing domain." Cell Growth Differ 6(12): 1495-503. Prag, S. and J. C. Adams (2003). "Molecular phylogeny of the kelch-repeat superfamily reveals an expansion of BTB/kelch proteins in animals." BMC Bioinformatics 4(1): 42. Xu, J., T. He, et al. (2004). "Molecular cloning and characterization of a novel human BTBD8 gene containing double BTB/POZ domains." Int J Mol Med 13(1): 193-7. |