 |
 |
 |
A specific subdomain for Carboxypeptidases A and Carboxypeptidases B.Carboxypeptidases (CP) are a subgroup of metallopeptidases, which remove amino acids from the C-termini of proteins and peptides by hydrolysis (Rawlings and Barrett 1995; Vendrell, Querol et al. 2000). knowledge on the structure and functionality of pancreatic carboxypeptidases has grown in a considerably way in the last few years (Aviles, Vendrell et al. 1993). If the cleavage mechanisms use an active site serine, cysteine or zinc, the group is referred to as metallocarboxypeptidases (Rawlings and Barrett 1995).. Altogether, there are 17 known members of the metallocarboxypeptidase gene family in most mammalian species investigated (Wei, Segura et al. 2002). All metallocarboxypeptidases can be grouped into one of two major subfamilies based primarily on amino acid sequences similarities. One group includes the digestive enzymes carboxypeptidase A (CPA) and carboxypeptidase B (CPB). The subdomain described here is characteristic for this protein group. Several non-mammal carboxypeptidase-like sequences have been reported in Swiss-prot and TrEMBL databases, so the cluster could be even bigger than 14 sequences as has been proposed (Reznik and Fricker 2001). In such way, the CPB and CPA carboxylases are closer groups to each other, than to any other group. The group is completed by CPU and other CPA-like hypothetical proteins, found in A. gambiae and other insects.The primary function of pancreatic CPA and B is to break peptides in the gut, following the action of chymotripsin and trypsin on ingested proteins. Similar to the pancreatic CPAs, MC-CPA also cleaves peptides with C-terminal aromatic/aliphatic residues. MC-CPA functions in the destruction of proteins and peptides by mast cells, presumably following the action of chymase. Additionally, Carboxypeptidase U (CPU) is a plasma enzyme that is produced in the liver and cleaves C-terminal lysines formed by the action of plasmin on fibrin (Reznik and Fricker 2001). The subdomain belongs to the already known domain Zinc peptidase (SM00235), but is not included in the Zinc-carboxypeptidase domain (PF00246), and is useful to distinguish between members of the different metallocarboxypeptidase families. Furthermore includes the active carboxypeptidase active site. The profile retrieves all the sequences that belong to this subfamily. Examples


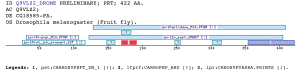
Taxonomic DistributionVertebrates: Canis familiaris, Homo sapiens, Sus scrofa . Bacteria: Streptomyces capreolus, Thermoactinomyces vulgaris. Invertebrates: Caenorhabditis briggsae, Caenorhabditis elegans, Lumbricus rubellus. Fungi: Metarhizium anisopliae, Neurospora crassa, Magnaporthe grisea. PSI-BLAST against ensembl (Apis mellifera)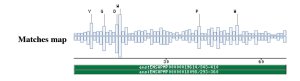
PSI-BLAST against ensembl (no taxonomic restriction)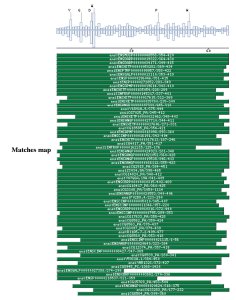
Comparison against UNIPROT
Retrived sequences in UNIPROT (txt format) Downloadable filesProfileMultiple Alignment PSI-BLAST against ensembl (Apis mellifera) PSI-BLAST against ensembl (no taxonomic restriction) REFERENCESAviles, F. X., J. Vendrell, et al. (1993). "Advances in metallo-procarboxypeptidases. Emerging details on the inhibition mechanism and on the activation process." Eur J Biochem 211(3): 381-9. Rawlings, N. D. and A. J. Barrett (1995). "Evolutionary families of metallopeptidases." Methods Enzymol 248: 183-228. Reznik, S. E. and L. D. Fricker (2001). "Carboxypeptidases from A to z: implications in embryonic development and Wnt binding." Cell Mol Life Sci 58(12-13): 1790-804. Vendrell, J., E. Querol, et al. (2000). "Metallocarboxypeptidases and their protein inhibitors. Structure, function and biomedical properties." Biochim Biophys Acta 1477(1-2): 284-98. Wei, S., S. Segura, et al. (2002). "Identification and characterization of three members of the human metallocarboxypeptidase gene family." J Biol Chem 277(17): 14954-64. |