 |
 |
 |
The Dynein Heavy Chain family caseDyneins have been implicated in a variety of microtubule-associated movements, including vesicle transport, cellular organization of membranous organelles, and flagellar motility among others (Oiwa and Sakakibara 2005) (Asai and Wilkes 2004) (Asai and Koonce 2001). The dynein ATPases are a family of motor enzymes that drive microtubule sliding in cilia and flagella and contribute to microtubule sliding cilia and flagella and contribute to microtubule-based transport inside cell (Mitchell and Brown 1994) (Porter, Knott et al. 1996). These enzymes convert the energy derived from nucleotide binding and hydrolysis into the directed movement of cellular cargoes toward the minus ends of microtubules. Axonemal dyneins can be divided in two groups: the outer dynein arms and the inner dynein arms, which have different functions in generating and propagating the flagellar waveforms. The outer dynein arms provide power to the flagellar beat. The Chlamydomonas sp. mutants lacking them swim with a reduced beat frequency (Brokaw and Kamiya 1987). In contrast the inner arms are essential for normal motility, as mutants with inner arm defects have near normal beat frequencies, but display aberrant waveforms.The Chlamydomonas outer arm is one of the most well characterized dynein complexes; it is composed of three dynein heavy chains (Dhc) 1 (ab and g), two intermediate chains, and several light chains (20 kD). The COOH-terminal and the central regions in the heavy chain of the outer arm are known because it forms a globular domain that interacts transiently with the microtubule doublet during the cross-bridge cycle. The more variable NH2-terminus is thought to form a flexible stem domain that extends to the base of the dynein arm and interacts with the isoform-specific intermediate chain and light chain (Myster, Knott et al. 1999). Most the efforts have been devoted to study how is the assembly of the subunits and what is the role of each subunit in the complex, specially in I1 Chlamydomonas reinhardtii complex, because it is now the best understood of all this kind complex. Furthermore, is another well known domain in the dynein protein family: the motor domain which contains several tandem linked AAA domains which form a ring (ATPases Associated with different cellular Activities) motif is present several times in the most of the matched proteins. This motif is defined by a conserved sequence of 230 to 250 amino acids that includes the Walker signature sequences of P-loop ATPases and other regions of similarity unique to AAA proteins (Sakato and King 2004). DYHC_AD5All the retrieved sequences by this profile are dynein heavy chains. All of them belong to two different dynein types: either the axonemal dyneins or the cytoplasmatic dyneins. The cytoplasmic dyneins are relatively simple because are composed of two identical dynein heavy chains (>500kDa) complexed with a cluster of intermediate (74 kDa) and light intermediate (50-55 kDa) chain components. On the other hand, the axonemal dyneins are much more diverse, and multiple dynein heavy chain isoforms have been detected in several species (Porter 1996). Not all the dynein heavy chains are matched with this domain, but it could means that the domain is specific for a dynein heavy chain protein subfamily, and would be a subfamily conformed either by axonemal and cytoplasmatic dynein heavy chains. In the specific case of dyneins is important to say, that the matched sequence and the profile, do not correspond to a coiled coil or a transmembrane region, because in this kind of proteins are many already known coiled-coil domains. It has been reported (King and Dutcher 1997) that a construct expressing the first 1249 amino acids of the Dynein 1-alpha heavy chain (Q9SMH3) but lacking the motor domain is able to assemble I1 complexes and target them to their correct location on the axoneme, although motility is not normal, in Chlamydomonas reinhardtii. This region includes the DYHC_AD domain. It means the DYHC_AD5 domain could be implied in the normal motility of the axoneme. Further experimentation is required to confirm it. This domain is very often found on the N-ter of the Pfam :Dynein_heavy (PF03028) domain located between AAA5 and AAA6. DYHC_AD1In this case the matched sequences, which confirm the cluster are dynein or dynein-like proteins. Within the cluster there is some hypothetical proteins too, which have not annotation about domains. The dynein sequences are only heavy chain sequences that fall in different classes: 1-beta DHC, DHC alpha, Ciliary DHC 7, Axonemal DHC 8 among others.The information obtained from this new domain could be useful to give some lights about the function of some sequences which its function is not known yet. Two well conserved patterns, that can be found close to the C-terminus of the domain: one of them could be defined as VhxRDV[I,V,L,F]xxL[I,A,V] and the other one is shorter but even better conserved and could be written as QLR[Y,F]Y. Is not ruled out that the two patterns could be merged, as happen in almost all the sequences in the cluster. These patterns are present in all these proteins, but do not define the cluster as well as the profile that retrieves the sequences, because the latter one is a more powerful tool to detect weak homologies, and skip problems like define one or two patterns. As happens in the DYHC_AD5 domain the retrieved sequences are dynein heavy chains, belonging to two different dynein types: axonemal dyneins or cytoplasmatic dyneins, so this domain could be another way to define another dynein subfamily, which would be conformed by axonemal and cytoplasmatic dyneins, because is not restricted to the distribution of this domain within the dynein heavy chains. Examples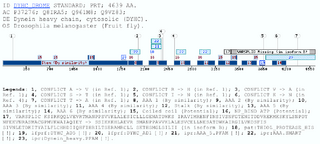



Taxonomic DistributionPlants: Arabidopsis thaliana, Nicotiana tabacum Insecta: Drosophila melanogaster, Anopheles gambiae Fungi: Emericella nidulans, Neurospora crassa, Cryptococcus neoformans Vertebrata: Homo sapiens, Mus musculus, Bos taurus Algae: Chlamydomonas reinhardtii Invertebrata: Anthocidaris crassispina, Tripneustes gratilla, Caenorhabditis elegans Protista: Trypanosoma brucei, Giardia lamblia, Leishmania major, Plasmodium falciparum
REFERENCESAsai, D. J. and M. P. Koonce (2001). "The dynein heavy chain: structure, mechanics and evolution." Trends Cell Biol 11(5): 196-202. Asai, D. J. and D. E. Wilkes (2004). "The dynein heavy chain family." J Eukaryot Microbiol 51(1): 23-9. Brokaw, C. J. and R. Kamiya (1987). "Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function." Cell Motil Cytoskeleton 8(1): 68-75. King, S. J. and S. K. Dutcher (1997). "Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains." J Cell Biol 136(1): 177-91. Mitchell, D. R. and K. S. Brown (1994). "Sequence analysis of the Chlamydomonas alpha and beta dynein heavy chain genes." J Cell Sci 107 (Pt 3): 635-44. Myster, S. H., J. A. Knott, et al. (1999). "Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas." J Cell Biol 146(4): 801-18. Oiwa, K. and H. Sakakibara (2005). "Recent progress in dynein structure and mechanism." Curr Opin Cell Biol 17(1): 98-103. Porter, M. E. (1996). "Axonemal dyneins: assembly, organization, and regulation." Curr Opin Cell Biol 8(1): 10-7. Porter, M. E., J. A. Knott, et al. (1996). "The dynein gene family in Chlamydomonas reinhardtii." Genetics 144(2): 569-85. Sakato, M. and S. M. King (2004). "Design and regulation of the AAA+ microtubule motor dynein." J Struct Biol 146(1-2): 58-71. |