Section |
Characteristics |
Example
|
| Description of the domain |
Provides information about the sequences retrieved by the profile, position of the matched region in representative proteins and other biochemical features of those proteins that have the new domain. |
|
| Examples |
The examples included in each domain page are graphics that are useful to analyze the domain composition of the proteins matched by the profile. They are also useful to give a graphical approach of the domain position in the protein. |
 |
| Taxonomic distribution |
This item enumerates some of the most representative groups, with some examples, that have the domain. |
|
| PSI-BLAST against ensembl |
A PSI-BLAST against the ensembl database (only the Apis mellifera proteome) has been performed with the multiple alignments as a starting point. The results in a graphic format are showed in this section, but the plain text file is also avilable in the Dowloadable files section. This is of particular importance for those domains that has been classified as Insect specific domains. The significant matches are displayed in green, and the non-significant are displayed in red. |
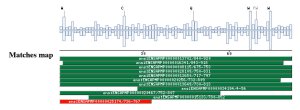 |
| PSI-BLAST against ensembl (no taxonomic restriction |
A PSI-BLAST against the ensembl database, with no taxonomic restrictions was done, to check what new organism were related with the profile and thus with the proposed domain. |
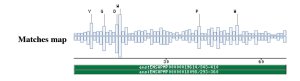 |
| Comparison against UNIPROT |
A search for related proteins was performed having as a starting point the multiple alignments. The retrieved sequences were examined to verify that the domain was insect specific or not. The retrieved sequences were included in the alignment to develop the final profile. |
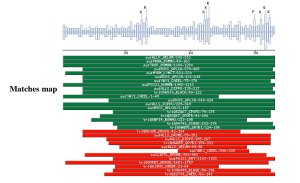 |
| Downloadable files |
This section includes all the result files in a plain text format. |
|


