- Con relación al alineamiento pareado cuales son la principales diferencias entre un alineamiento global y uno local y en casos usted recomendaría utilizar cada uno de ellos? De manera general, como se calcula el "score" de una alineamiento pareado?
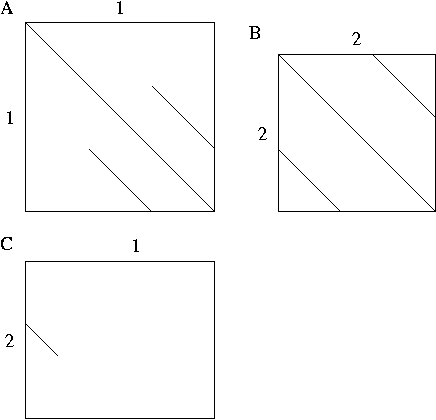
- Las figuras de abajo son dotplots obtenidos con 2 proteínas hipotéticas
"1" and "2". La figura A representa la proteína "1" comparada contra ella
misma, La figura B representa la proteína "2" comparada contra ella misma,
y la figura C representa la proteína "1" alineada con la proteína "2".
- Porque usted obtiene una diagonal larga en la figuras A y B? (2 pts)
- En las mismas figuras, que representan las diagonales pequeñas? (2 pts)
- Trate de deducir los posibles dominios presentes en estas dos proteínas
usando las 3 figuras (A,B,C) y haga una representación esquemática de
ellos. (6 pts)
- Debajo encontrará la salida de una búsqueda BLAST usando SwissProt y TrEMBL
como bases de datos. Este resultado fue obtenido usando el programa
blastp (protein query vs protein database) y la proteína UFD1_YEAST como
query.
- Que representan las "X" en la secuencia query? (vea el primer alineamiento)
- Cuantas proteínas en SwissProt tienen un "score" que pueda ser considerado como significativo en esta búsqueda?
- Cual es la primera proteína con un "score" que pueda ser considerado como significativo?
- Cual es la matriz de peso utilizada para esta búsqueda y porque?Si pudiera escoger la matriz cual seleccionaria y porque?
- Usted desea realizar un estudio en mamíferos utilizando la siguiente secuencia de proteína que obtuvo experimentalmente con el fin de conocer su función, estructura y evolución.Para ello utilizará las herramientas bioinformáticas disponibles y que ha conocido durante el curso.
EDHVKLVNEVTEFAKKCAADESAENCDKSLHTLFGDKLCTVATLRATYGELADCCEKQEP ERNECFLTHKDDHPNLPKLKPEPDAQCAAFQEDPDKFLGKYLYEVARRHPYFYGPELLFH AEEYKADFTECCPADDKLACLIPKLDALKERILLSSAKERLKCSSFQNFGERAVKAWSVA RLSQKFPKADFAEVSKIVTDLTKVHKECCHGDLLECADDRADLAKYICEHQDSISGKLKA CCDKPLLQKSHCIAEVKEDDLPSDLPALAADFAEDKEICKHYKDAKDVFLGTFLYEYSRR HPDYSVSLLLRIAKTYEATLEKCCAEADPPACYRTVFDQFTPLVEEPKSLVKKNCDLFEE VGEYDFQNALIVRYTKKAPQVSTPTLVEIGRTLGKVGSRCCKLPESERL
Determine:- Su función y la familia a la que pertenece
- Cual es Secuencia del gen correspondiente?
- Cuantas proteinas en SwissProt tienen un "score" que pueda ser considerado como significativo en esta búsqueda?
- Cual es la matriz de peso utilizada por defecto para la búsqueda y porque?
- Identifique en los resultados de su búsqueda un grupo de secuencias potencialmente homólogas (mínimo 8) a su query. Recuperelas, reportelas y realice un alinemiento multiple con ellas (reportelo), que le indica el alineamiento?
BLAST2.0 query receipt
Thanks. Your request has been filed with the following data:
Program: blastp Database: swisstrembl Number: 4979 e-mail: Format: SwissProt_ID Sequence: ufd1_yeast
Processing, please wait... blastall.remote -s blast.cscs.ch -p blastp -d swisstrembl wwwtmp/sq.4979.txt -M blosum62
-K 0 -e 10.0 -F T -G def -E def -X def -v 50 -b 50 -g T |
Here are your search results: SIB BLAST network server version 1.3 of November 15, 2000 Welcome to the SIB BLAST Network Service (julier) ============================================================================== Swiss Institute of Bioinformatics (SIB) Ludwig Institute for Cancer Research (LICR) Swiss Institute for Experimental Cancer Research (ISREC) ============================================================================== If results of this search are reported or published, please mention that the computation was performed at the SIB using the BLAST network service. PEPTIDE SEQUENCE DATABASES swiss SwissProt + updates 26-Jan-2001 swisstrembl SwissProt+Trembl+TrUpdates 26-Jan-2001 nr Non-redundant SP+Trembl+TrUpdates+gp+gpu 26-Jan-2001 gpupdate cumulative weekly updates of gp, currently 26-Jan-2001 shuffled SwissProt r30 shuffled in windows of 20 aa yeast S.cerevisiae translated genome ORFs NUCLEOTIDE SEQUENCE DATABASES embre * EMBL Data Library, Release 65, December 2000 embu * EmblUpdate 31-Jan-2001 embl * EMBL+EmblUpdate 31-Jan-2001 EST Database of Expressed Sequence Tags 31-Jan-2001 GSS Genome Survey Sequences 31-Jan-2001 STS Database of Sequence Tagged Sites 31-Jan-2001 nr * Non-redundant EMBL+EmblUpdate+GenBank 31-Jan-2001 repbase + Repbase, February 1996 simple Simple sequence repeats (J. Mol. Evol. 40:120, 1995) yeast S.cerevisiae genome worm C.elegans genome * The est, gss, and sts divisions of EMBL are excluded from these databases. + Contains sequences from the files humrep.ref, invrep.ref, mamrep.ref, plnrep.ref, rodrep.ref, and vertrep.ref from repbase. ============================================================================ T he BLAST Network Service uses a server developed at SIB and the NCBI BLAST 2 software. For problems regarding this site, please contact one of the following persons: - problems with the programs: Christian Iseli (chris@cmpteam4.unil.ch) - problems with the databases: Victor Jongeneel (Victor.Jongeneel@isb-sib.ch) ============================================================================ BL ASTP 2.1.2 [Oct-19-2000] Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= sp|P53044|UFD1_YEAST (UFD1..)UBIQUITIN FUSION DEGR (361 letters) Database: swiss_nr; swiss_varsplic_nr; trembl_new_nr; trembl_nr; trembl_varsplic_nr 500,285 sequences; 166,584,592 total letters Searching..................................................done
Score E Sequences producing significant alignments: (bits) Value sp|P53044|UFD1_YEAST (UFD1..)UBIQUITIN FUSION DEGRADATION PROTE... 655 0.0 tr|AJ005825|O42915 (UFD1..)UBIQUITIN FUSION DEGRADATION PROTEIN... 264 1e-69 tn|AF234601|AAG27535 UFD1.[Rattus norvegicus] 204 1e-51 sp_vs|Q92890-01|Q92890 SHORT ISOFORM OF Q92890[Homo sapiens] 203 2e-51 sp|P70362|UFD1_MOUSE (UFD1L)UBIQUITIN FUSION DEGRADATION PROTEI... 201 9e-51 tn|AF233524|AAG25922 Ubiquitin fusion degradation 1-like protei... 200 2e-50 sp|Q9VTF9|UFD1_DROME (CG6233)UBIQUITIN FUSION DEGRADATION PROTE... 196 4e-49 tr|AC006841|Q9SJV0 (AT2G21270)PUTATIVE UBIQUITIN FUSION-DEGRADA... 192 3e-48 tr|AL035679|Q9SVK0 (F19H22.30..)PUTATIVE UBIQUITIN-DEPENDENT PR... 190 2e-47 sp|Q92890|UFD1_HUMAN (UFD1L)UBIQUITIN FUSION DEGRADATION PROTEI... 185 4e-46 sp|Q19584|UFD1_CAEEL (F19B6.2)UBIQUITIN FUSION DEGRADATION PROT... 170 2e-41 tr|AC005315|O81075 (T9I4.15)PUTATIVE UBIQUITIN.[Arabidopsis tha... 169 3e-41 tr|Z69635|Q9U3I6 (F19B6.2B)F19B6.2B PROTEIN.[Caenorhabditis ele... 169 3e-41 tr|AF083031|Q9SEV9 (UFD1)UBIQUITIN FUSION DEGRADATION PROTEIN.[... 106 4e-22 tr|Z97338|O23395 (AT4G15420)SIMILAR TO UFD1 PROTEIN (UFD1 LIKE ... 102 6e-21 tn|AF315735|AAG31651 PRLI-interacting factor K (Fragment).[Arab... 102 6e-21 tr|AE001179|O51745 (BB0805)POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSF... 35 1.4 tr|AC010793|Q9MA08 F20B17.10.[Arabidopsis thaliana] 34 3.2 tr|AC010924|Q9S9M2 (T24D18.23)T24D18.23 PROTEIN.[Arabidopsis th... 34 3.2 tr|Z97182|O07779 (RV0599C..)HYPOTHETICAL 8.2 KDA PROTEIN.[Mycob... 34 3.2 tr|AC013289|Q9SFH8 (T6C23.7)HYPOTHETICAL 88.6 KDA PROTEIN.[Arab... 33 4.1 tr|AC010924|Q9S9M6 (T24D18.19)T24D18.19 PROTEIN.[Arabidopsis th... 33 5.4 tr|AC010924|Q9S9M4 (T24D18.21)T24D18.21 PROTEIN.[Arabidopsis th... 32 7.1 tr|AF020904|O16859 (MAFP3E)AGGREGATION FACTOR PROTEIN 3, FORM E... 32 9.3 tr|AB037819|Q9P2E9 (KIAA1398)KIAA1398 PROTEIN (FRAGMENT).[Homo ... 32 9.3 tr|AE004191|Q9KSZ6 (VC1110)SERYL-TRNA SYNTHETASE.[Vibrio cholerae] 32 9.3 tn|AL132765|CAC10588 (RRBP1)BA462D18.3.2 (ribosome binding prot... 32 9.3 >sp|P53044|UFD1_YEAST (UFD1..)UBIQUITIN FUSION DEGRADATION PROTEIN 1 (UB FUSION PROTEIN 1) (POLYMERASE-INTERACTING PROTEIN 3).[Saccharomyces cerevisiae] Length = 361 Score = 655 bits (1672), Expect = 0.0 Identities = 329/347 (94%), Positives = 329/347 (94%) Query: 15 VNMPQTFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTAN 74 VNMPQTFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTAN Sbjct: 15 VNMPQTFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTAN 74 Query: 75 ETGRVTHGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDF 134 ETGRVTHGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDF Sbjct: 75 ETGRVTHGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDF 134 Query: 135 LDISDPKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVT 194 LDISDPKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVT Sbjct: 135 LDISDPKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVT 194 Query: 195 DFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNK 254 DFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNK Sbjct: 195 DFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNK 254 Query: 255 LSKFVGQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEES 314 LSKFVGQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEES Sbjct: 255 LSKFVGQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEES 314 Query: 315 AAGSKSSEQNFQGQGISLRKSNXXXXXXXXXXXXXXXXXXPEVIEID 361 AAGSKSSEQNFQGQGISLRKSN PEVIEID Sbjct: 315 AAGSKSSEQNFQGQGISLRKSNKRKTKSDHDSSKSKAPKSPEVIEID 361 >tr|AJ005825|O42915 (UFD1..)UBIQUITIN FUSION DEGRADATION PROTEIN 1 (UB FUSION PROTEIN 1).[Schizosaccharomyces pombe] Length = 342 Score = 264 bits (667), Expect = 1e-69 Identities = 145/320 (45%), Positives = 197/320 (61%), Gaps = 24/320 (7%) Query: 16 NMPQTFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANE 75 N+ Q F+ +RCYP+AM+ R + N+GGK+ LPPSAL KLS LN+ YPMLF Sbjct: 27 NVNQRFDTRYRCYPVAMIPGEERPN-VNYGGKVILPPSALEKLSRLNVSYPMLFDFENEA 85 Query: 76 TGRVTHGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFL 135 + THGGVLEFIAEEGRVYLP WMM TL ++PG L+++ +TD+ G +VKL+PQSV+FL Sbjct: 86 AEKKTHGGVLEFIAEEGRVYLPYWMMTTLSLEPGDLVRVINTDIAQGSYVKLQPQSVNFL 145 Query: 136 DISDPKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTD 195 DI+D +AVLEN LRNFSTLT D+ EI YN + ++IK+++V+P+ S + V+ETDLV D Sbjct: 146 DITDHRAVLENALRNFSTLTKSDIFEILYNDQVYQIKVIDVQPDDSRHVVSVVETDLVVD 205 Query: 196 FAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNKL 255 F PP+GY E + Q +++N G QG+M T+I Y + + L Sbjct: 206 FDPPIGYEE-------SLQKNKQQNIAGV----------QGTMVTKIGYDELVRQGDSNL 248 Query: 256 SKFVGQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEESA 315 K G G ++GK PK ++ D + PA L LP G FFG+P P +E+S Sbjct: 249 MK--GTGTKLNGKEVAEVPKINLLD--VEKQECPAPLILPLGTYFFGYPYKAPSIEEDSK 304 Query: 316 AGSKSSEQNFQGQGISLRKS 335 + F+G G SLR S Sbjct: 305 KDPNLFK--FEGAGTSLRAS 322 >tn|AF234601|AAG27535 UFD1.[Rattus norvegicus] Length = 307 Score = 204 bits (514), Expect = 1e-51 Identities = 128/320 (40%), Positives = 179/320 (55%), Gaps = 42/320 (13%) Query: 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80 F +RC+ ++M+ + D GGKI +PPSAL +LS LNI YPMLFKLT + R+T Sbjct: 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78 Query: 81 HGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDP 140 H GVLEF+A+EG YLP WMM+ L ++ G L+Q+ S ++ + + K +PQS DFLDI++P Sbjct: 79 HCGVLEFVADEGICYLPHWMMQNLLLEEGGLVQVESVNLQVATYSKFQPQSPDFLDITNP 138 Query: 141 KAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPV 200 KAVLEN LRNF+ LT DVI I+YN K ++++++E KP+ K++ +IE D+ DF P+ Sbjct: 139 KAVLENALRNFACLTTGDVIAINYNEKIYELRVMETKPD---KAVSIIECDMNVDFDAPL 195 Query: 201 GYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNKLSKFVG 260 GY EP+ + Q +E S+ D++G A F G Sbjct: 196 GYKEPE----RPVQHEE-------------------SIEGEADHSGYAGEV--GFRAFSG 230 Query: 261 QGQNISGKAPKAEPK-QDIK--DMKITFDGEPAKLDLPEGQLFF---GFPMVLPKEDEES 314 G + GK EP IK D+K KL G++ F PMV K+ EE Sbjct: 231 SGNRLDGKKKGVEPSPSPIKPGDIKRGIPNYEFKL----GKITFIRNSRPMV--KKVEED 284 Query: 315 AAGSKSSEQNFQGQGISLRK 334 AG + F G+G SLRK Sbjct: 285 EAGGRFVA--FSGEGQSLRK 302 >sp_vs|Q92890-01|Q92890 SHORT ISOFORM OF Q92890[Homo sapiens] Length = 307 Score = 203 bits (512), Expect = 2e-51 Identities = 122/320 (38%), Positives = 178/320 (55%), Gaps = 42/320 (13%) Query: 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80 F +RC+ ++M+ + D GGKI +PPSAL +LS LNI YPMLFKLT + R+T Sbjct: 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78 Query: 81 HGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDP 140 H GVLEF+A+EG YLP WMM+ L ++ G L+Q+ S ++ + + K +PQS DFLDI++P Sbjct: 79 HCGVLEFVADEGICYLPHWMMQNLLLEEGGLVQVESVNLQVATYSKFQPQSPDFLDITNP 138 Query: 141 KAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPV 200 KAVLEN LRNF+ LT DVI I+YN K ++++++E KP+ K++ +IE D+ DF P+ Sbjct: 139 KAVLENALRNFACLTTGDVIAINYNEKIYELRVMETKPD---KAVSIIECDMNVDFDAPL 195 Query: 201 GYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNKLSKFVG 260 GY EP+ +Q + ++++ G+ D++G A F G Sbjct: 196 GYKEPE------RQVQHEESTEGEA-----------------DHSGYAGEL--GFRAFSG 230 Query: 261 QGQNISGKAPKAEPKQ------DIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEES 314 G + GK EP DIK ++ + K+ F L K+ EE Sbjct: 231 SGNRLDGKKKGVEPSPSPIKPGDIKRGIPNYEFKLGKI------TFIRNSRPLVKKVEED 284 Query: 315 AAGSKSSEQNFQGQGISLRK 334 AG + F G+G SLRK Sbjct: 285 EAGGRFVA--FSGEGQSLRK 302 >sp|P70362|UFD1_MOUSE (UFD1L)UBIQUITIN FUSION DEGRADATION PROTEIN 1 HOMOLOG (UB FUSION PROTEIN 1).[Mus musculus] Length = 307 Score = 201 bits (506), Expect = 9e-51 Identities = 122/320 (38%), Positives = 174/320 (54%), Gaps = 42/320 (13%) Query: 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80 F +RC+ ++M+ + D GGKI +PPSAL +LS LNI YPMLFKLT + R+T Sbjct: 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78 Query: 81 HGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDP 140 H GVLEF+A+EG YLP WMM+ L ++ G L+Q+ S ++ + + K +PQS DFLDI++P Sbjct: 79 HCGVLEFVADEGICYLPHWMMQNLLLEEGGLVQVESVNLQVATYSKFQPQSPDFLDITNP 138 Query: 141 KAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPV 200 KAVLEN LRNF+ +T DVI I+YN K ++++++E KP+ K++ +IE D+ DF P+ Sbjct: 139 KAVLENALRNFACMTTGDVIAINYNEKIYELRVMETKPD---KAVSIIECDMNVDFDAPL 195 Query: 201 GYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNKLSKFVG 260 GY EP + Q +E S+ D++G A F G Sbjct: 196 GYKEPK----RPVQHEE-------------------SIEGEADHSGYAGEV--GFRAFSG 230 Query: 261 QGQNISGKAPKAEPKQ------DIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEES 314 G + GK EP DIK ++ + K+ F L K+ EE Sbjct: 231 SGNRLDGKKKGVEPSPSPIKPGDIKRGIPNYEFKLGKI------TFIRNSRPLVKKVEED 284 Query: 315 AAGSKSSEQNFQGQGISLRK 334 AG + F G+G SLRK Sbjct: 285 EAGGRFVA--FSGEGQSLRK 302 >tn|AF233524|AAG25922 Ubiquitin fusion degradation 1-like protein (Fragment).[Xenopus laevis] Length = 305 Score = 200 bits (504), Expect = 2e-50 Identities = 116/286 (40%), Positives = 160/286 (55%), Gaps = 18/286 (6%) Query: 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80 F +RCY ++M+ + D GGKI +PPSAL +LS LNI YPMLFKLT + R+T Sbjct: 19 FSTQYRCYSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78 Query: 81 HGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDP 140 H GVLEF+A+EG YLP WMM+ L ++ LLQ+ S ++ + + K +PQS DFLDI++P Sbjct: 79 HCGVLEFVADEGVCYLPHWMMQNLLLEEEGLLQVESVNLQVATYSKFQPQSPDFLDITNP 138 Query: 141 KAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPV 200 KAVLEN LRNF+ LT DVI I+YN K ++++++E KP+ K++ +IE D+ DF P+ Sbjct: 139 KAVLENALRNFACLTTGDVIAINYNEKIYELRVMETKPD---KAVSIIECDMNVDFDAPL 195 Query: 201 GYVEPDYKALKAQQ------------DKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIA 248 GY EPD + +Q D + G G LD G G + + GI Sbjct: 196 GYKEPDRHTQQEEQTEIEPDHSEYAADLGFRAFSGSGNRLDGKKKGIGPSPSPLKPEGIR 255 Query: 249 NSSRNKLSKFVGQGQNISGKAP--KAEPKQDIKDMKITFDGEPAKL 292 N K +G I P K + D + I F GE L Sbjct: 256 RGIPNYDFK-IGTITFIRNSRPQTKKVEEDDTANQFIAFSGEGQSL 300 >sp|Q9VTF9|UFD1_DROME (CG6233)UBIQUITIN FUSION DEGRADATION PROTEIN 1 HOMOLOG (UB FUSION PROTEIN 1).[Drosophila melanogaster] Length = 316 Score = 196 bits (492), Expect = 4e-49 Identities = 120/324 (37%), Positives = 176/324 (54%), Gaps = 31/324 (9%) Query: 19 QTFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGR 78 + F ++C+ ++M+ R D GGKI +PPSAL L+ LN+ YPMLFKLT + R Sbjct: 15 RNFHANYKCFSVSMLPGNERTD-VEKGGKIIMPPSALDTLTRLNVEYPMLFKLTNVKKSR 73 Query: 79 VTHGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDIS 138 +H GVLEF+A+EG+ YLP WMME L + G +L I S +P+ F K +P S DFLDI+ Sbjct: 74 SSHAGVLEFVADEGKCYLPHWMMENLLLGEGDILNIESVSLPVATFSKFQPHSTDFLDIT 133 Query: 139 DPKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAP 198 +PKAVLEN LRNF+ LT DVI I YN K +++ +LE KP ++ +IE D+ +F Sbjct: 134 NPKAVLENALRNFACLTRGDVIAIKYNKKVYELCVLETKP---GNAVSIIECDMNVEFEA 190 Query: 199 PVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMST-RIDYAGIANSSRNK-LS 256 PVGY +D + + G GQ QG+ T + AG N+ + + Sbjct: 191 PVGY-----------KDHSETQASGSGQ--------QGAAGTVGGEIAGATNAILEEVVE 231 Query: 257 KFVGQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFG---FPMVLPKEDEE 313 F G G + GK K + K+ G P D G + F P+ ++++ Sbjct: 232 TFKGSGVRLDGKKKKESQLETPVVKKVLARGVP-DYDFQFGLIRFDRNIRPISDRSQEDD 290 Query: 314 SAAGS--KSSEQNFQGQGISLRKS 335 + AG+ S ++F G G S++K+ Sbjct: 291 AVAGNADASDAESFHGTGFSMKKT 314 >tr|AC006841|Q9SJV0 (AT2G21270)PUTATIVE UBIQUITIN FUSION-DEGRADATION PROTEIN.[Arabidopsis thaliana] Length = 320 Score = 192 bits (484), Expect = 3e-48 Identities = 124/310 (40%), Positives = 173/310 (55%), Gaps = 36/310 (11%) Query: 20 TFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRV 79 TFE+ +RCYP + ++ K G KI +PPSAL +L+ L+I YPMLF+L RV Sbjct: 13 TFEQSYRCYPASFID----KPQLESGDKIIMPPSALDRLASLHIDYPMLFELRNAGIERV 68 Query: 80 THGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISD 139 TH GVLEFIAEEG +Y+P WMM+ L +Q G ++++ + +P G +VKL+P + DFLDIS+ Sbjct: 69 THCGVLEFIAEEGMIYMPYWMMQNLLLQEGDIVRVRNVTLPKGTYVKLQPHTTDFLDISN 128 Query: 140 PKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPP 199 PKA+LE LRN+S LT D I + YN K + I I+E KP + +I +IETD DFAPP Sbjct: 129 PKAILETALRNYSCLTSGDSIMVPYNNKKYFIDIVETKP---ANAISIIETDCEVDFAPP 185 Query: 200 VGYVEPD-----------YKALKAQQDKEKK-NSF-GKGQVLD---------PSVLGQGS 237 + Y EP+ KA + + E K N F G G+ LD P+ +G Sbjct: 186 LDYKEPERPTAPSAAKGPAKAEEVVDEPEPKFNPFTGSGRRLDGRPLAYEPAPASSSKGK 245 Query: 238 MSTRIDYAGIANSSRNKLSKFVGQGQNISG----KAPK-AEPKQDIKDMKITFDGEPAKL 292 + G ++ + + QG+ + G +APK A PK + K T E K Sbjct: 246 QPVVANGNGQSSVASSSEKATRAQGKLVFGANGNRAPKEAAPK--VGAAKETKKEEQEKK 303 Query: 293 DLPEGQLFFG 302 D P+ Q F G Sbjct: 304 DEPKFQAFSG 313 >tr|AL035679|Q9SVK0 (F19H22.30..)PUTATIVE UBIQUITIN-DEPENDENT PROTEOLYTIC PROTEIN.[Arabidopsis thaliana] Length = 315 Score = 190 bits (478), Expect = 2e-47 Identities = 104/256 (40%), Positives = 145/256 (56%), Gaps = 30/256 (11%) Query: 20 TFEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRV 79 TFE+ +RCYP + ++ K G KI +PPSAL +L+ L I YPMLF+L T Sbjct: 12 TFEQTYRCYPSSFID----KPQIESGDKIIMPPSALDRLASLQIDYPMLFELRNASTDSF 67 Query: 80 THGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISD 139 +H GVLEFIAEEG +Y+P WMM+ L +Q G ++++ + +P G +VKL+P + DFLDI++ Sbjct: 68 SHCGVLEFIAEEGVIYIPYWMMQNLLLQEGDMVRVRNVTLPKGTYVKLQPHTTDFLDIAN 127 Query: 140 PKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPP 199 PKA+LE LRN+S LTV D I + YN K + I I+E KP S I +IETD DFAPP Sbjct: 128 PKAILETALRNYSCLTVGDSIMVPYNNKKYFIDIVEAKP---SNGISIIETDCEVDFAPP 184 Query: 200 VGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLGQGSMSTRIDYAGIANSSRNKLSKFV 259 + Y EP+ A E K + +D A + K + F Sbjct: 185 LDYKEPERPVAPAPAKGEAK-------------------AKEVDVA----EAEPKFNPFT 221 Query: 260 GQGQNISGKAPKAEPK 275 G G+ + G+ EP+ Sbjct: 222 GSGRRLDGRPLSYEPQ 237 >sp|Q92890|UFD1_HUMAN (UFD1L)UBIQUITIN FUSION DEGRADATION PROTEIN 1 HOMOLOG (UB FUSION PROTEIN 1).[Homo sapiens] Length = 343 Score = 185 bits (466), Expect = 4e-46 Identities = 124/356 (34%), Positives = 179/356 (49%), Gaps = 78/356 (21%) Query: 21 FEEFFRCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVT 80 F +RC+ ++M+ + D GGKI +PPSAL +LS LNI YPMLFKLT + R+T Sbjct: 19 FSTQYRCFSVSMLAGPNDRSDVEKGGKIIMPPSALDQLSRLNITYPMLFKLTNKNSDRMT 78 Query: 81 HGGVLEFIAEEGRVYLPQWMM-------------ETLGIQ-------------------- 107 H GVLEF+A+EG YLP WMM ET+ +Q Sbjct: 79 HCGVLEFVADEGICYLPHWMMQNLLLEEDGLVQLETVNLQVATYSKSKFCYLPHWMMQNL 138 Query: 108 ---PGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLENVLRNFSTLTVDDVIEISY 164 G L+Q+ S ++ + + K +PQS DFLDI++PKAVLEN LRNF+ LT DVI I+Y Sbjct: 139 LLEEGGLVQVESVNLQVATYSKFQPQSPDFLDITNPKAVLENALRNFACLTTGDVIAINY 198 Query: 165 NGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFGK 224 N K ++++++E KP+ K++ +IE D+ DF P+GY EP+ +Q + ++++ G+ Sbjct: 199 NEKIYELRVMETKPD---KAVSIIECDMNVDFDAPLGYKEPE------RQVQHEESTEGE 249 Query: 225 GQVLDPSVLGQGSMSTRIDYAGIANSSRNKLSKFVGQGQNISGKAPKAEPKQ------DI 278 D++G A F G G + GK EP DI Sbjct: 250 A-----------------DHSGYAGEL--GFRAFSGSGNRLDGKKKGVEPSPSPIKPGDI 290 Query: 279 KDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEESAAGSKSSEQNFQGQGISLRK 334 K ++ + K+ F L K+ EE AG + F G+G SLRK Sbjct: 291 KRGIPNYEFKLGKI------TFIRNSRPLVKKVEEDEAGGRFVA--FSGEGQSLRK 338 >sp|Q19584|UFD1_CAEEL (F19B6.2)UBIQUITIN FUSION DEGRADATION PROTEIN 1 HOMOLOG (UB FUSION PROTEIN 1).[Caenorhabditis elegans] Length = 342 Score = 170 bits (427), Expect = 2e-41 Identities = 123/327 (37%), Positives = 174/327 (52%), Gaps = 22/327 (6%) Query: 21 FEEFFRCY-PIAMMNDRIRK-DDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGR 78 +++ F Y P+ + N K + N+GGKI LP SAL+ L NI PMLFKLT R Sbjct: 21 YDQTFVVYGPVFLPNATQSKISEINYGGKILLPSSALNLLMQYNIPMPMLFKLTNMAVQR 80 Query: 79 VTHGGVLEFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDIS 138 VTH GVLEF A EG+ LP WMM+ LG+ G ++I S +P F KL+P S++FL+I+ Sbjct: 81 VTHCGVLEFSAPEGQAILPLWMMQQLGLDDGDTIRIESATLPKATFAKLKPMSLEFLNIT 140 Query: 139 DPKAVLENVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAP 198 +PKAVLE LR ++ LT +D I SY G+T + ++++KP + S+C+IE D+ DF P Sbjct: 141 NPKAVLEVELRKYACLTKNDRIPTSYAGQTLEFLVVDLKP---ANSVCIIECDVNLDFDP 197 Query: 199 PVGYVEPDYK---ALKAQQDKEKKNSF-GKGQVLDPSVLGQGSMSTRI-DYAGIANSSRN 253 P GYVE + A+ A+ ++F G GQ S G G +T + AG + Sbjct: 198 PEGYVEQPRQVTPAVTAKPPAPDASAFIGAGQKAGGSG-GTGQNATSVFGGAGRRLDGKK 256 Query: 254 KLSKFV----GQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPM--VL 307 K S V G G + S AP A I + + D +P ++ L + + VL Sbjct: 257 KPSSSVSLSDGTGVSTSNAAPVANDLPAIPPVVVNEDYKPGRVSF----LRYDYKRVDVL 312 Query: 308 PKEDEESAAGSKSSEQN-FQGQGISLR 333 KE E A N F+G +LR Sbjct: 313 EKELREKEASKAGQPSNVFRGGNRTLR 339 >tr|AC005315|O81075 (T9I4.15)PUTATIVE UBIQUITIN.[Arabidopsis thaliana] Length = 292 Score = 169 bits (425), Expect = 3e-41 Identities = 99/263 (37%), Positives = 144/263 (54%), Gaps = 31/263 (11%) Query: 47 KIFLPPSALSKLSMLNIRYPMLFKLTANETGRVTHGGVLEFIAEEGRVYLPQWMMETLGI 106 + +PPSAL +L+ L+I YPMLF+L+ + +H GVLEF A+EG VYLP WMM+ + + Sbjct: 10 QFIMPPSALDRLASLHIEYPMLFQLSNVSVEKTSHCGVLEFTADEGLVYLPYWMMQNMSL 69 Query: 107 QPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLENVLRNFSTLTVDDVIEISYNG 166 + G ++Q+ + + G ++KL+P + DFLDIS+PKA+LE LR++S LT D I + YN Sbjct: 70 EEGDVMQVKNISLVKGTYIKLQPHTQDFLDISNPKAILETTLRSYSCLTTGDTIMVPYNN 129 Query: 167 KTFKIKILEVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQ 226 K + I ++E KP S ++ +IETD DFAPP+ Y EP+ K Q Sbjct: 130 KQYYINVVEAKP---SSAVSIIETDCEVDFAPPLDYKEPE-----------------KPQ 169 Query: 227 VLDPSVLGQGSMSTRIDYAGIANSSRNKLSKFVGQGQNISGKA-PKAEP---KQDIKDMK 282 L PS ++ S K + F G G+ + GKA + EP KQ K + Sbjct: 170 KLTPS----NKRPLQVKEEEEPASKVPKFTPFTGSGKRLDGKAQTQTEPEDTKQQEKPTE 225 Query: 283 ITFDGEPAKLDLP---EGQLFFG 302 D E + P G+L FG Sbjct: 226 NGKDDEKLSVTTPRQISGKLVFG 248 >tr|Z69635|Q9U3I6 (F19B6.2B)F19B6.2B PROTEIN.[Caenorhabditis elegans] Length = 336 Score = 169 bits (425), Expect = 3e-41 Identities = 118/305 (38%), Positives = 164/305 (53%), Gaps = 20/305 (6%) Query: 41 DANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVTHGGVLEFIAEEGRVYLPQWM 100 + N+GGKI LP SAL+ L NI PMLFKLT RVTH GVLEF A EG+ LP WM Sbjct: 37 EINYGGKILLPSSALNLLMQYNIPMPMLFKLTNMAVQRVTHCGVLEFSAPEGQAILPLWM 96 Query: 101 METLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLENVLRNFSTLTVDDVI 160 M+ LG+ G ++I S +P F KL+P S++FL+I++PKAVLE LR ++ LT +D I Sbjct: 97 MQQLGLDDGDTIRIESATLPKATFAKLKPMSLEFLNITNPKAVLEVELRKYACLTKNDRI 156 Query: 161 EISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPVGYVEPDYK---ALKAQQDKE 217 SY G+T + ++++KP + S+C+IE D+ DF PP GYVE + A+ A+ Sbjct: 157 PTSYAGQTLEFLVVDLKP---ANSVCIIECDVNLDFDPPEGYVEQPRQVTPAVTAKPPAP 213 Query: 218 KKNSF-GKGQVLDPSVLGQGSMSTRI-DYAGIANSSRNKLSKFV----GQGQNISGKAPK 271 ++F G GQ S G G +T + AG + K S V G G + S AP Sbjct: 214 DASAFIGAGQKAGGSG-GTGQNATSVFGGAGRRLDGKKKPSSSVSLSDGTGVSTSNAAPV 272 Query: 272 AEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPM--VLPKEDEESAAGSKSSEQN-FQGQ 328 A I + + D +P ++ L + + VL KE E A N F+G Sbjct: 273 ANDLPAIPPVVVNEDYKPGRVSF----LRYDYKRVDVLEKELREKEASKAGQPSNVFRGG 328 Query: 329 GISLR 333 +LR Sbjct: 329 NRTLR 333 >tr|AF083031|Q9SEV9 (UFD1)UBIQUITIN FUSION DEGRADATION PROTEIN.[Guillardia theta] Length = 175 Score = 106 bits (262), Expect = 4e-22 Identities = 60/167 (35%), Positives = 94/167 (55%), Gaps = 7/167 (4%) Query: 26 RCYPIAMMNDRIRKDDANFGGKIFLPPSALSKLSMLNIRYPMLFKLTANETGRVTHGGVL 85 + YP++ I K G KI LP S L+ L+ + P++F++ + + H GV Sbjct: 9 KTYPLSF----IGKSFLENGDKIVLPQSILNYLNQNDDLNPIIFEILNLDNNKKCHCGVY 64 Query: 86 EFIAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLE 145 EF +++G Y+P WM + L I GS L + G F+K++PQ +F IS+PKA+LE Sbjct: 65 EFTSDDGCAYIPYWMFKNLEINEGSPLCFIQKCLEKGYFLKIQPQQKEFFQISNPKAILE 124 Query: 146 NVLRNFSTLTVDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDL 192 LR +++LT + I I YN + + I+EVKP +I +I+TDL Sbjct: 125 LNLRKYTSLTKKNTISIEYNNNIYWLNIVEVKP---GNAINIIDTDL 168 >tr|Z97338|O23395 (AT4G15420)SIMILAR TO UFD1 PROTEIN (UFD1 LIKE PROTEIN).[Arabidopsis thaliana] Length = 778 Score = 102 bits (252), Expect = 6e-21 Identities = 65/191 (34%), Positives = 107/191 (55%), Gaps = 18/191 (9%) Query: 45 GGKIFLPPSALSKLSMLNI--RYPMLFKLTA---NETGRVTHGGVLEFIAEEGRVYLPQ- 98 G KI LPPS ++LS + P+ F+L+ + + TH GVLEF AE+G + LP Sbjct: 308 GDKIKLPPSCFTELSDQGAFDKGPLYFELSVVDHADNKKTTHSGVLEFTAEDGTIGLPPH 367 Query: 99 -W--MMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLENVLRNFSTLT 155 W + T L++I +P G + KL+P ++ F D+ + KA+LE +LR +TL+ Sbjct: 368 VWSNLFSTHDPMDVPLVEIRYIRLPKGSYAKLQPDNLGFSDLPNHKAILETILRQHATLS 427 Query: 156 VDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQD 215 +DDV+ ++Y ++K+++LE++P + SI V+ETD+ D V PD + + Q Sbjct: 428 LDDVLLVNYGQVSYKLQVLELRP---ATSISVLETDIEVDI------VSPDIVSDQPNQH 478 Query: 216 KEKKNSFGKGQ 226 K +GK + Sbjct: 479 VLKPLQYGKSE 489 >tn|AF315735|AAG31651 PRLI-interacting factor K (Fragment).[Arabidopsis thaliana] Length = 574 Score = 102 bits (252), Expect = 6e-21 Identities = 65/191 (34%), Positives = 107/191 (55%), Gaps = 18/191 (9%) Query: 45 GGKIFLPPSALSKLSMLNI--RYPMLFKLTA---NETGRVTHGGVLEFIAEEGRVYLPQ- 98 G KI LPPS ++LS + P+ F+L+ + + TH GVLEF AE+G + LP Sbjct: 104 GDKIKLPPSCFTELSDQGAFDKGPLYFELSVVDHADNKKTTHSGVLEFTAEDGTIGLPPH 163 Query: 99 -W--MMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDISDPKAVLENVLRNFSTLT 155 W + T L++I +P G + KL+P ++ F D+ + KA+LE +LR +TL+ Sbjct: 164 VWSNLFSTHDPMDVPLVEIRYIRLPKGSYAKLQPDNLGFSDLPNHKAILETILRQHATLS 223 Query: 156 VDDVIEISYNGKTFKIKILEVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQD 215 +DDV+ ++Y ++K+++LE++P + SI V+ETD+ D V PD + + Q Sbjct: 224 LDDVLLVNYGQVSYKLQVLELRP---ATSISVLETDIEVDI------VSPDIVSDQPNQH 274 Query: 216 KEKKNSFGKGQ 226 K +GK + Sbjct: 275 VLKPLQYGKSE 285 >tr|AE001179|O51745 (BB0805)POLYRIBONUCLEOTIDE NUCLEOTIDYLTRANSFERASE (PNPA).[Borrelia burgdorferi] Length = 722 Score = 34.8 bits (78), Expect = 1.4 Identities = 32/88 (36%), Positives = 44/88 (49%), Gaps = 13/88 (14%) Query: 217 EKKNSFGKGQVLDPSVLGQGSMSTRID-----YAGIANSSRNKLSKFVGQGQNISGKA-- 269 +K NSFG L P+ +G +STR+ Y + N+ S+F G G+NI G A Sbjct: 630 KKINSFGAFIELTPAK--EGFLSTRLKPRDSKYGSGRFGNSNRYSRFGGGGENIRGNAGL 687 Query: 270 ---PKAEPKQDIKDMKITFDGEPAKLDL 294 PK E Q IK +KI + K+DL Sbjct: 688 VRPPKLEEGQRIK-VKIIDIDKFGKIDL 714 >tr|AC010793|Q9MA08 F20B17.10.[Arabidopsis thaliana] Length = 1487 Score = 33.6 bits (75), Expect = 3.2 Identities = 19/65 (29%), Positives = 36/65 (55%), Gaps = 2/65 (3%) Query: 175 EVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLG 234 +V +S+++ V T L T + VGY++P+Y ++ Q +K + + G VL + G Sbjct: 574 KVSDFGTSRTVTVDHTHLTTVVSGTVGYMDPEY--FQSSQFTDKSDVYSFGVVLAELITG 631 Query: 235 QGSMS 239 + S+S Sbjct: 632 EKSVS 636 >tr|AC010924|Q9S9M2 (T24D18.23)T24D18.23 PROTEIN.[Arabidopsis thaliana] Length = 700 Score = 33.6 bits (75), Expect = 3.2 Identities = 17/50 (34%), Positives = 26/50 (52%) Query: 174 LEVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFG 223 ++V +S+S+ + +T L T A GYV+P+Y DK SFG Sbjct: 502 VKVSDFGTSRSVTIDQTHLTTQVAGTFGYVDPEYFQSSKFTDKSDVYSFG 551 >tr|Z97182|O07779 (RV0599C..)HYPOTHETICAL 8.2 KDA PROTEIN.[Mycobacterium tuberculosis] Length = 78 Score = 33.6 bits (75), Expect = 3.2 Identities = 16/45 (35%), Positives = 28/45 (61%), Gaps = 5/45 (11%) Query: 92 GRVYLPQWMMETLGIQPGSLLQISSTD-----VPLGQFVKLEPQS 131 GR+ +P+ + E LG+QPGS ++IS +P G+ +LE ++ Sbjct: 9 GRIVVPKPLREALGLQPGSTVEISRYGAGLHLIPTGRTARLEEEN 53 >tr|AC013289|Q9SFH8 (T6C23.7)HYPOTHETICAL 88.6 KDA PROTEIN.[Arabidopsis thaliana] Length = 792 Score = 33.2 bits (74), Expect = 4.1 Identities = 19/65 (29%), Positives = 36/65 (55%), Gaps = 2/65 (3%) Query: 175 EVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLG 234 +V +S+++ V T L T + VGY++P+Y ++ Q +K + + G VL + G Sbjct: 588 KVSDFGTSRTVTVDHTHLTTVVSGTVGYMDPEY--FQSSQFTDKSDVYSFGVVLVELITG 645 Query: 235 QGSMS 239 + S+S Sbjct: 646 EKSIS 650 >tr|AC010924|Q9S9M6 (T24D18.19)T24D18.19 PROTEIN.[Arabidopsis thaliana] Length = 1016 Score = 32.8 bits (73), Expect = 5.4 Identities = 21/67 (31%), Positives = 33/67 (48%), Gaps = 2/67 (2%) Query: 175 EVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLG 234 +V +S+SI + +T L T A GY++P+Y DK SF G VL + G Sbjct: 948 KVSDFGTSRSITIAQTHLTTLVAGTFGYMDPEYFLSSQYTDKSDVYSF--GVVLVELITG 1005 Query: 235 QGSMSTR 241 + +S + Sbjct: 1006 EKPLSRK 1012 >tr|AC010924|Q9S9M4 (T24D18.21)T24D18.21 PROTEIN.[Arabidopsis thaliana] Length = 664 Score = 32.5 bits (72), Expect = 7.1 Identities = 18/61 (29%), Positives = 33/61 (53%), Gaps = 2/61 (3%) Query: 175 EVKPESSSKSICVIETDLVTDFAPPVGYVEPDYKALKAQQDKEKKNSFGKGQVLDPSVLG 234 +V +S+S+ + +T L T A GYV+P+Y ++ + EK + + G VL + G Sbjct: 472 KVSDFGTSRSVTIDQTHLTTQVAGTFGYVDPEY--FQSSKFTEKSDVYSFGVVLVELLTG 529 Query: 235 Q 235 + Sbjct: 530 E 530 >tr|AF020904|O16859 (MAFP3E)AGGREGATION F ACTOR PROTEIN 3, FORM E (FRAGMENT).[Microciona prolifera] Length = 825 Score = 32.1 bits (71), Expect = 9.3 Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 11/91 (12%) Query: 88 IAEEGRVYLPQWMMETLGIQPGSLLQISSTDVPLGQFVKLEPQSVDFLDIS-----DPKA 142 +AEEG Y P + T+ I P S ++ ++++ L LE + L I+ D + Sbjct: 329 MAEEGEDYDPANVPSTVTIAPNSSMECFTSNLVLDDGESLEEDEIVVLSIAAVDPDDSRI 388 Query: 143 VLENVLRNFSTLTVDD----VIEISYNGKTF 169 ++ NV N + + +DD VI + + T+ Sbjct: 389 IVSNV--NVTVVIIDDDELQVITVQFEQSTY 417 >tr|AB037819|Q9P2E9 (KIAA1398)KIAA1398 PROTEIN (FRAGMENT).[Homo sapiens] Length = 1456 Score = 32.1 bits (71), Expect = 9.3 Identities = 22/76 (28%), Positives = 33/76 (42%) Query: 260 GQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEESAAGSK 319 G+G I GK + Q K IT G+ A+ EG+ G P K D + G K Sbjct: 785 GEGAPIQGKKADSVANQGTKVEGITNQGKKAEGSPSEGKKAEGSPNQGKKADAAANQGKK 844 Query: 320 SSEQNFQGQGISLRKS 335 + + QG+ + +S Sbjct: 845 TESASVQGRNTDVAQS 860 >tr|AE004191|Q9KSZ6 (VC1110)SERYL-TRNA SYNTHETASE.[Vibrio cholerae] Length = 435 Score = 32.1 bits (71), Expect = 9.3 Identities = 29/124 (23%), Positives = 50/124 (39%), Gaps = 5/124 (4%) Query: 206 DYKALKAQQDKEKKNSFGKGQVLDPSV---LGQGSMSTRIDYAGIANSSRNKLSKFVGQG 262 D K L+ + D+ +G LD L + S +++ + S+RN +SK +GQ Sbjct: 3 DSKLLRTELDETAAKLARRGFKLDVETIRTLEEQRKSIQVEVENL-QSTRNSISKQIGQL 61 Query: 263 QNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEESAAGSKSSE 322 KA KQ I + D + +LD QL +P +++ K Sbjct: 62 MASGDKAGAEAVKQQIGTLGDDLDAKKVELDAVMAQL-DAITQTVPNIPDDAVPNGKDDS 120 Query: 323 QNFQ 326 +N + Sbjct: 121 ENVE 124 >tn|AL132765|CAC10588 (RRBP1)BA462D18.3.2 (ribosome binding protein 1 (dog 180 kDa homolog)) (Fragment).[Homo sapiens] Length = 1208 Score = 32.1 bits (71), Expect = 9.3 Identities = 22/76 (28%), Positives = 33/76 (42%) Query: 260 GQGQNISGKAPKAEPKQDIKDMKITFDGEPAKLDLPEGQLFFGFPMVLPKEDEESAAGSK 319 G+G I GK + Q K IT G+ A+ EG+ G P K D + G K Sbjct: 536 GEGAPIQGKKADSVANQGTKVEGITNQGKKAEGSPSEGKKAEGSPNQGKKADAAANQGKK 595 Query: 320 SSEQNFQGQGISLRKS 335 + + QG+ + +S Sbjct: 596 TESASVQGRNTDVAQS 611 Database: swiss_nr Posted date: Jan 26, 2001 6:11 AM Number of letters in database: 33,669,062 Number of sequences in database: 92,100 Database: swiss_varsplic_nr Posted date: Jan 26, 2001 6:11 AM Number of letters in database: 1,587,947 Number of sequences in database: 2435 Database: trembl_new_nr Posted date: Jan 26, 2001 6:12 AM Number of letters in database: 15,364,861 Number of sequences in database: 46,874 Database: trembl_nr Posted date: Jan 26, 2001 6:16 AM Number of letters in database: 115,571,020 Number of sequences in database: 358,354 Database: trembl_varsplic_nr Posted date: Jan 26, 2001 6:17 AM Number of letters in database: 391,702 Number of sequences in database: 522 Lambda K H 0.315 0.136 0.382 Gapped Lambda K H 0.270 0.0470 0.230 Matrix: BLOSUM62 Gap Penalties: Existence: 11, Extension: 1 Number of Hits to DB: 107471254 Number of Sequences: 500285 Number of extensions: 4473755 Number of successful extensions: 7695 Number of sequences better than 10.0: 27 N umber of HSP's better than 10.0 without gapping: 20 Number of HSP's successfully gapped in prelim test: 7 Number of HSP's that attempted gapping in prelim test: 7650 Number of HSP's gapped (non-prelim): 32 length of query: 361 length of database: 166,584,592 effective HSP length: 57 effective length of query: 304 effective length of database: 138,068,347 effective search space: 41972777488 effective search space used: 41972777488 T: 11 A: 40 X1: 16 ( 7.3 bits) X2: 38 (14.8 bits) X3: 64 (24.9 bits) S1: 42 (22.0 bits) S2: 71 (32.1 bits) Thank you for using BLAST2.0