Grail2 predicts a number of exons, but no information about first/last exons.
The following output has been obtained using Grail2 with the options Shadow Exons, Translation, PolyA Sites, and CpG Islands.
[grail2exons -> Exons] St Fr Start End ORFstart ORFend Score Quality 1- f 2 3024 3188 2991 3188 92.000 excellent 2- f 1 4350 4457 4310 4468 100.000 excellent 3- f 0 5113 5216 5086 5355 92.000 excellent 4- f 0 5458 5496 5440 5586 91.000 excellent 5- f 1 6947 7053 6923 7057 100.000 excellent 6- f 1 7310 7472 7217 7498 72.000 good 7- f 1 7578 7644 7568 7648 98.000 excellent 8- f 0 7737 7938 7672 8043 95.000 excellent 9- f 0 8044 8103 8044 8103 68.000 good 10- f 1 9071 9287 9059 9364 98.000 excellent 11- f 1 10080 10186 9938 10186 72.000 good 12- f 2 15468 15578 15447 15695 70.000 good 13- f 0 18439 18616 18256 18639 97.000 excellent 14- f 1 19685 19836 19604 19840 83.000 excellent 15- f 1 19878 19999 19841 19999 94.000 excellent 16- r 2 19437 19562 19254 19622 75.000 excellent 17- r 1 8123 8278 7241 8278 78.000 excellent 18- r 1 7704 7840 7241 8278 83.000 excellent[grail2exons -> Shadow Exons] St Fr Start End ORFstart ORFend Score Quality 19- r 2 17854 17932 17757 18041 68.000 good 20- r 2 17414 17497 17355 17543 50.000 good 21- r 1 13406 13510 13313 13510 54.000 good 22- r 1 11586 11678 11561 11686 51.000 good 23- r 1 10733 10807 10634 10807 59.000 good 24- r 2 3547 3639 3507 3662 57.000 good[grail2exons -> Exon Translations] 25- EWTEAKELLQEEEEEEEEEDILSRDPSPEPPSHKLQRVQEKAGKPRRVRVREEL 26- LNEEAWFVVLTTGSSVTLLFWLLARLLNKAKIMTT 27- EPELPLDLCTMGEMEQLRQEAEQLKKQIAVTPEP 28- DARKACADITLA 29- LVSGLEVVGRVQMRTRRTLRGHLAKIYAMHWATDS 30- VHAIPLRSSWVMTCAYAPSGNFVACGGLDNMCSIYNLKSREGNVKVSRELSAHT 31- YLSCCRFLDDNNIVTSSGDTT 32- ALWDIETGQQKTVFVGHTGDCMSLAVSPDYKLFISGACDASAKLWDVREGTCRQTFTGHESDINAI 33- ESFKANDKESRVCFAQIHC 34- FFPNGEAICTGSDDASCRLFDLRADQELTAYSQESIICGITSVAFSLSGRLLFAGYDDFNCNVWDSLKCERV 35- ILSGHDNRVSCLGVTADGMAVATGSWDSFLKIWN 36- MAELSEEALLSVLPTIRVPKAGDRVHKDECAFSFDT 37- ESEGGLYICMNTFLGFGKQYVERHFNKTGQRVYLHLRRTRRPVGTVAGVEHPDTVHKEL 38- KEEDTSAGTGDPPRKKPTRLAIGELSARRTLSLMSDLRALEGTNWRHDDK 39- VEGGFDLTEDKFEFDEDVKIVILPDYLEIARDGLGGLPD 40- MYICVRHAFLVPLRRGWKKMLDPLERESEVVVNVDVLQKSS 41- GKRPSALSENVKDLKEGVVLGTGRFLKAGGGAREPNQDHDKENQHFALLES 42- SKRSRRKANSKVLGRSPLTILQDDNSPGTLTLRQVKEEGGENCH[PolyA Sites]Str Start End Score f 9701 9706 1.00 f 12612 12617 0.65[CPG Islands] Start End CpGscore GCscore 14251 14474 0.62 60.28 15232 15655 0.90 61.36
Click here for more informartion on the output format.
MZEF predict internal coding exons. The prediction for each strand is done separately.
The following output has been optained using the strand 1.
MZEF Results - human - unknownStrand = 1
Overlap = 0
Prior Prob. = .02
Internal coding exons predicted by MZEF
Sequence_length: 20000 G+C_content: 0.514
| Coordinates | P | Fr1 | Fr2 | Fr3 | Orf | 3ss | Cds | 5ss |
|---|---|---|---|---|---|---|---|---|
| 5458 - 5496 | 0.958 | 0.600 | 0.520 | 0.402 | 112 | 0.577 | 0.558 | 0.626 |
| 6947 - 7053 | 0.567 | 0.430 | 0.567 | 0.349 | 212 | 0.541 | 0.548 | 0.524 |
| 7310 - 7472 | 0.818 | 0.464 | 0.576 | 0.380 | 112 | 0.529 | 0.543 | 0.619 |
| 7578 - 7644 | 0.857 | 0.337 | 0.603 | 0.391 | 212 | 0.517 | 0.539 | 0.591 |
| 9071 - 9287 | 0.991 | 0.389 | 0.607 | 0.346 | 212 | 0.484 | 0.525 | 0.612 |
| 18439 - 18564 | 0.997 | 0.628 | 0.418 | 0.427 | 122 | 0.556 | 0.561 | 0.615 |
| 19685 - 19751 | 0.700 | 0.465 | 0.641 | 0.465 | 111 | 0.521 | 0.589 | 0.545 |
HMMgene returns a rich output. It predicts first/last exons when possible.
HMMgene finds 4 open reading frames (2 optimals and 2 sub-optimals), when the search is performed activating the option 2 best prediction.
The option Predict signals returns all the signal founds in the sequences, which can help to validate a prediction.
Explanation of output format## gff-version 1## date: Thu Aug 29 11:42:34 2002## HMMgene1.1a (human model sim10gc.C.bsmod)# SEQ: unknown 20000 (+) A:4886 C:5118 G:5156 T:4840emb|AC002397|AC002397 HMMgene1.1a firstex 1545 1586 0.647 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_1 3024 3170 0.653 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_2 5113 5199 0.504 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_3 5458 5496 0.991 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_4 6947 7053 0.998 + 2 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_5 7133 7196 0.996 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_6 7310 7472 0.993 + 1 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_7 7578 7644 0.998 + 2 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_8 7737 7938 0.993 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_9 9071 9287 0.975 + 1 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a lastex 10080 10186 0.969 + 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a CDS 1545 10186 0.187 + . bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a firstex 15468 15578 1.000 + 0 bestparse:cds_2emb|AC002397|AC002397 HMMgene1.1a exon_1 18439 18564 0.970 + 0 bestparse:cds_2emb|AC002397|AC002397 HMMgene1.1a exon_2 19685 19751 0.610 + 1 bestparse:cds_2emb|AC002397|AC002397 HMMgene1.1a exon_3 19878 19977 0.613 + 2 bestparse:cds_2emb|AC002397|AC002397 HMMgene1.1a CDS 15468 20000 0.549 + . bestparse:cds_2emb|AC002397|AC002397 HMMgene1.1a firstex 1545 1586 0.647 + 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_1 3024 3170 0.653 + 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_2 5458 5496 0.991 + 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_3 6947 7053 0.998 + 2 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_4 7133 7196 0.996 + 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_5 7310 7472 0.993 + 1 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_6 7578 7644 0.998 + 2 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_7 7737 7938 0.993 + 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_8 9071 9287 0.975 + 1 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a lastex 10080 10186 0.969 + 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a CDS 1545 10186 0.159 + . subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a firstex 15468 15578 1.000 + 0 subopt_1:cds_2emb|AC002397|AC002397 HMMgene1.1a exon_1 18439 18564 0.970 + 0 subopt_1:cds_2emb|AC002397|AC002397 HMMgene1.1a exon_2 19685 19751 0.610 + 1 subopt_1:cds_2emb|AC002397|AC002397 HMMgene1.1a exon_3 19878 19977 0.613 + 2 subopt_1:cds_2emb|AC002397|AC002397 HMMgene1.1a CDS 15468 20000 0.549 + . subopt_1:cds_2emb|AC002397|AC002397 HMMgene1.1a ACC 20 21 0.002 + 0emb|AC002397|AC002397 HMMgene1.1a START 60 62 0.103 + .emb|AC002397|AC002397 HMMgene1.1a DON 62 63 0.105 + 0emb|AC002397|AC002397 HMMgene1.1a START 417 419 0.003 + .emb|AC002397|AC002397 HMMgene1.1a DON 427 428 0.003 + 2emb|AC002397|AC002397 HMMgene1.1a START 540 542 0.002 + .emb|AC002397|AC002397 HMMgene1.1a START 576 578 0.019 + .emb|AC002397|AC002397 HMMgene1.1a DON 653 654 0.024 + 0emb|AC002397|AC002397 HMMgene1.1a START 986 988 0.089 + .emb|AC002397|AC002397 HMMgene1.1a DON 1027 1028 0.088 + 0emb|AC002397|AC002397 HMMgene1.1a START 1178 1180 0.077 + .emb|AC002397|AC002397 HMMgene1.1a DON 1180 1181 0.077 + 0emb|AC002397|AC002397 HMMgene1.1a START 1545 1547 0.649 + .emb|AC002397|AC002397 HMMgene1.1a DON 1586 1587 0.647 + 0emb|AC002397|AC002397 HMMgene1.1a START 2177 2179 0.002 + .emb|AC002397|AC002397 HMMgene1.1a DON 2179 2180 0.003 + 0emb|AC002397|AC002397 HMMgene1.1a START 2284 2286 0.004 + .emb|AC002397|AC002397 HMMgene1.1a DON 2364 2365 0.004 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 2524 2525 0.004 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 2556 2557 0.005 + 2emb|AC002397|AC002397 HMMgene1.1a ACC 2650 2651 0.095 + 0emb|AC002397|AC002397 HMMgene1.1a DON 2656 2657 0.002 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 2656 2657 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a DON 2791 2792 0.104 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 3023 3024 0.939 + 0emb|AC002397|AC002397 HMMgene1.1a DON 3134 3135 0.005 + 0emb|AC002397|AC002397 HMMgene1.1a DON 3164 3165 0.222 + 0emb|AC002397|AC002397 HMMgene1.1a DON 3170 3171 0.653 + 0emb|AC002397|AC002397 HMMgene1.1a DON 3184 3185 0.008 + 2emb|AC002397|AC002397 HMMgene1.1a STOP 3186 3188 0.051 + .emb|AC002397|AC002397 HMMgene1.1a ACC 4242 4243 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a DON 4263 4264 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 4349 4350 0.007 + 2emb|AC002397|AC002397 HMMgene1.1a START 4366 4368 0.003 + .emb|AC002397|AC002397 HMMgene1.1a DON 4368 4369 0.009 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 4687 4688 0.011 + 0emb|AC002397|AC002397 HMMgene1.1a DON 4795 4796 0.002 + 0emb|AC002397|AC002397 HMMgene1.1a DON 4816 4817 0.009 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 5112 5113 0.504 + 0emb|AC002397|AC002397 HMMgene1.1a START 5143 5145 0.060 + .emb|AC002397|AC002397 HMMgene1.1a START 5152 5154 0.007 + .emb|AC002397|AC002397 HMMgene1.1a DON 5199 5200 0.571 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 5209 5210 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 5457 5458 0.991 + 0emb|AC002397|AC002397 HMMgene1.1a DON 5496 5497 0.991 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 6946 6947 0.998 + 0emb|AC002397|AC002397 HMMgene1.1a DON 7053 7054 0.999 + 2emb|AC002397|AC002397 HMMgene1.1a ACC 7132 7133 0.999 + 2emb|AC002397|AC002397 HMMgene1.1a DON 7196 7197 0.996 + 0emb|AC002397|AC002397 HMMgene1.1a DON 7253 7254 0.003 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 7270 7271 0.007 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 7309 7310 0.993 + 0emb|AC002397|AC002397 HMMgene1.1a DON 7472 7473 1.000 + 1emb|AC002397|AC002397 HMMgene1.1a ACC 7498 7499 0.004 + 1emb|AC002397|AC002397 HMMgene1.1a DON 7552 7553 0.004 + 1emb|AC002397|AC002397 HMMgene1.1a ACC 7577 7578 0.998 + 1emb|AC002397|AC002397 HMMgene1.1a DON 7644 7645 0.998 + 2emb|AC002397|AC002397 HMMgene1.1a ACC 7736 7737 0.999 + 2emb|AC002397|AC002397 HMMgene1.1a ACC 7822 7823 0.002 + 1emb|AC002397|AC002397 HMMgene1.1a DON 7875 7876 0.005 + 0emb|AC002397|AC002397 HMMgene1.1a DON 7938 7939 0.995 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 8043 8044 0.004 + 0emb|AC002397|AC002397 HMMgene1.1a STOP 8101 8103 0.003 + .emb|AC002397|AC002397 HMMgene1.1a START 8852 8854 0.004 + .emb|AC002397|AC002397 HMMgene1.1a DON 8902 8903 0.004 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 9070 9071 0.997 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 9169 9170 0.003 + 0emb|AC002397|AC002397 HMMgene1.1a DON 9287 9288 0.978 + 1emb|AC002397|AC002397 HMMgene1.1a DON 9293 9294 0.022 + 1emb|AC002397|AC002397 HMMgene1.1a ACC 9334 9335 0.003 + 1emb|AC002397|AC002397 HMMgene1.1a STOP 9415 9417 0.003 + .emb|AC002397|AC002397 HMMgene1.1a ACC 9474 9475 0.001 + 1emb|AC002397|AC002397 HMMgene1.1a STOP 9492 9494 0.002 + .emb|AC002397|AC002397 HMMgene1.1a ACC 9992 9993 0.015 + 1emb|AC002397|AC002397 HMMgene1.1a ACC 10031 10032 0.007 + 1emb|AC002397|AC002397 HMMgene1.1a ACC 10079 10080 0.969 + 1emb|AC002397|AC002397 HMMgene1.1a DON 10151 10152 0.001 + 1emb|AC002397|AC002397 HMMgene1.1a STOP 10184 10186 0.990 + .emb|AC002397|AC002397 HMMgene1.1a ACC 14290 14291 0.004 + 1emb|AC002397|AC002397 HMMgene1.1a STOP 14431 14433 0.004 + .emb|AC002397|AC002397 HMMgene1.1a START 15468 15470 1.000 + .emb|AC002397|AC002397 HMMgene1.1a DON 15578 15579 1.000 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 15932 15933 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a DON 16013 16014 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 18108 18109 0.045 + 0emb|AC002397|AC002397 HMMgene1.1a DON 18165 18166 0.045 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 18438 18439 0.996 + 0emb|AC002397|AC002397 HMMgene1.1a DON 18564 18565 0.971 + 0emb|AC002397|AC002397 HMMgene1.1a DON 18573 18574 0.007 + 0emb|AC002397|AC002397 HMMgene1.1a DON 18582 18583 0.015 + 0emb|AC002397|AC002397 HMMgene1.1a DON 18600 18601 0.001 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 19603 19604 0.005 + 0emb|AC002397|AC002397 HMMgene1.1a ACC 19684 19685 0.994 + 0emb|AC002397|AC002397 HMMgene1.1a DON 19751 19752 0.615 + 1emb|AC002397|AC002397 HMMgene1.1a DON 19787 19788 0.016 + 1emb|AC002397|AC002397 HMMgene1.1a DON 19836 19837 0.001 + 2emb|AC002397|AC002397 HMMgene1.1a STOP 19838 19840 0.367 + .emb|AC002397|AC002397 HMMgene1.1a ACC 19877 19878 0.629 + 1emb|AC002397|AC002397 HMMgene1.1a ACC 19877 19878 0.001 + 2emb|AC002397|AC002397 HMMgene1.1a STOP 19882 19884 0.001 + .emb|AC002397|AC002397 HMMgene1.1a DON 19930 19931 0.016 + 0emb|AC002397|AC002397 HMMgene1.1a DON 19977 19978 0.613 + 2# SEQ: emb|AC002397|AC002397 20000 (-) A:4840 C:5156 G:5118 T:4886emb|AC002397|AC002397 HMMgene1.1a firstex 15508 15623 0.699 - 2 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_1 14273 14499 0.252 - 1 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_2 12424 12717 0.497 - 1 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_3 12194 12297 0.908 - 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a lastex 11723 11878 0.283 - 0 bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a CDS 11723 15623 0.029 - . bestparse:cds_1emb|AC002397|AC002397 HMMgene1.1a firstex 15508 15623 0.699 - 2 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_1 14264 14499 0.251 - 1 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_2 12424 12717 0.497 - 1 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a exon_3 12194 12297 0.908 - 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a lastex 11723 11878 0.283 - 0 subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a CDS 11723 15623 0.029 - . subopt_1:cds_1emb|AC002397|AC002397 HMMgene1.1a START 19996 19998 0.010 - .emb|AC002397|AC002397 HMMgene1.1a DON 19957 19958 0.010 - 2emb|AC002397|AC002397 HMMgene1.1a START 19942 19944 0.101 - .emb|AC002397|AC002397 HMMgene1.1a DON 19902 19903 0.099 - 0emb|AC002397|AC002397 HMMgene1.1a DON 19897 19898 0.002 - 2emb|AC002397|AC002397 HMMgene1.1a START 19747 19749 0.019 - .emb|AC002397|AC002397 HMMgene1.1a DON 19737 19738 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a DON 19721 19722 0.016 - 1emb|AC002397|AC002397 HMMgene1.1a DON 19669 19670 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a START 18971 18973 0.005 - .emb|AC002397|AC002397 HMMgene1.1a ACC 18950 18951 0.047 - 0emb|AC002397|AC002397 HMMgene1.1a DON 18940 18941 0.005 - 0emb|AC002397|AC002397 HMMgene1.1a START 18915 18917 0.031 - .emb|AC002397|AC002397 HMMgene1.1a ACC 18844 18845 0.006 - 2emb|AC002397|AC002397 HMMgene1.1a DON 18821 18822 0.077 - 0emb|AC002397|AC002397 HMMgene1.1a DON 18821 18822 0.006 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 18577 18578 0.002 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 18477 18478 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a DON 18426 18427 0.004 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 18136 18137 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 18110 18111 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 18050 18051 0.036 - 0emb|AC002397|AC002397 HMMgene1.1a DON 17998 17999 0.038 - 1emb|AC002397|AC002397 HMMgene1.1a START 17188 17190 0.001 - .emb|AC002397|AC002397 HMMgene1.1a DON 17187 17188 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a START 16986 16988 0.003 - .emb|AC002397|AC002397 HMMgene1.1a DON 16974 16975 0.003 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 16875 16876 0.002 - 2emb|AC002397|AC002397 HMMgene1.1a DON 16828 16829 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 16458 16459 0.003 - 0emb|AC002397|AC002397 HMMgene1.1a START 16425 16427 0.001 - .emb|AC002397|AC002397 HMMgene1.1a DON 16361 16362 0.002 - 1ggemb|AC002397|AC002397 HMMgene1.1a ACC 16054 16055 0.012 - 0emb|AC002397|AC002397 HMMgene1.1a DON 15923 15924 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a DON 15867 15868 0.010 - 1emb|AC002397|AC002397 HMMgene1.1a START 15825 15827 0.013 - .emb|AC002397|AC002397 HMMgene1.1a DON 15786 15787 0.011 - 2emb|AC002397|AC002397 HMMgene1.1a DON 15768 15769 0.002 - 2emb|AC002397|AC002397 HMMgene1.1a START 15726 15728 0.004 - .emb|AC002397|AC002397 HMMgene1.1a DON 15707 15708 0.004 - 0emb|AC002397|AC002397 HMMgene1.1a START 15633 15635 0.041 - .emb|AC002397|AC002397 HMMgene1.1a ACC 15627 15628 0.004 - 1emb|AC002397|AC002397 HMMgene1.1a START 15621 15623 0.700 - .emb|AC002397|AC002397 HMMgene1.1a START 15543 15545 0.009 - .emb|AC002397|AC002397 HMMgene1.1a DON 15507 15508 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a DON 15507 15508 0.749 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 15419 15420 0.094 - 0emb|AC002397|AC002397 HMMgene1.1a DON 15375 15376 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a DON 15337 15338 0.026 - 1emb|AC002397|AC002397 HMMgene1.1a DON 15264 15265 0.066 - 2emb|AC002397|AC002397 HMMgene1.1a START 14775 14777 0.007 - .emb|AC002397|AC002397 HMMgene1.1a ACC 14770 14771 0.005 - 0emb|AC002397|AC002397 HMMgene1.1a START 14753 14755 0.025 - .emb|AC002397|AC002397 HMMgene1.1a ACC 14735 14736 0.008 - 2emb|AC002397|AC002397 HMMgene1.1a DON 14707 14708 0.008 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 14684 14685 0.004 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 14622 14623 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 14607 14608 0.005 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 14607 14608 0.008 - 2emb|AC002397|AC002397 HMMgene1.1a DON 14589 14590 0.049 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14589 14590 0.009 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 14542 14543 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 14499 14500 0.061 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 14499 14500 0.820 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 14484 14485 0.002 - 2emb|AC002397|AC002397 HMMgene1.1a DON 14445 14446 0.056 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14445 14446 0.036 - 2emb|AC002397|AC002397 HMMgene1.1a DON 14441 14442 0.013 - 0emb|AC002397|AC002397 HMMgene1.1a DON 14434 14435 0.066 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14427 14428 0.004 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14411 14412 0.146 - 0emb|AC002397|AC002397 HMMgene1.1a DON 14411 14412 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 14360 14361 0.007 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 14343 14344 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14338 14339 0.039 - 1emb|AC002397|AC002397 HMMgene1.1a START 14330 14332 0.005 - .emb|AC002397|AC002397 HMMgene1.1a DON 14328 14329 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a DON 14314 14315 0.015 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14272 14273 0.254 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14263 14264 0.253 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14241 14242 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a DON 14241 14242 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a ACC 14240 14241 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 14240 14241 0.004 - 2emb|AC002397|AC002397 HMMgene1.1a DON 14212 14213 0.018 - 0emb|AC002397|AC002397 HMMgene1.1a DON 14212 14213 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 13852 13853 0.013 - 2emb|AC002397|AC002397 HMMgene1.1a DON 13775 13776 0.013 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 13666 13667 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 13501 13502 0.175 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 13501 13502 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a DON 13455 13456 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 13444 13445 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a DON 13383 13384 0.048 - 1emb|AC002397|AC002397 HMMgene1.1a DON 13380 13381 0.114 - 1emb|AC002397|AC002397 HMMgene1.1a DON 13370 13371 0.012 - 2emb|AC002397|AC002397 HMMgene1.1a DON 13354 13355 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 13159 13160 0.033 - 2emb|AC002397|AC002397 HMMgene1.1a DON 13115 13116 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a DON 13109 13110 0.031 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 12946 12947 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 12946 12947 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a DON 12914 12915 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a DON 12828 12829 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 12717 12718 0.989 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 12642 12643 0.010 - 1emb|AC002397|AC002397 HMMgene1.1a DON 12423 12424 0.501 - 1emb|AC002397|AC002397 HMMgene1.1a DON 12355 12356 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a DON 12348 12349 0.483 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 12297 12298 0.970 - 1emb|AC002397|AC002397 HMMgene1.1a DON 12262 12263 0.007 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 12229 12230 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a DON 12193 12194 0.920 - 0emb|AC002397|AC002397 HMMgene1.1a DON 12177 12178 0.054 - 1emb|AC002397|AC002397 HMMgene1.1a DON 12160 12161 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 11918 11919 0.004 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 11899 11900 0.030 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 11878 11879 0.334 - 0emb|AC002397|AC002397 HMMgene1.1a DON 11877 11878 0.003 - 0emb|AC002397|AC002397 HMMgene1.1a DON 11877 11878 0.014 - 1emb|AC002397|AC002397 HMMgene1.1a DON 11856 11857 0.034 - 1emb|AC002397|AC002397 HMMgene1.1a DON 11848 11849 0.005 - 0emb|AC002397|AC002397 HMMgene1.1a DON 11827 11828 0.008 - 0emb|AC002397|AC002397 HMMgene1.1a DON 11735 11736 0.008 - 2emb|AC002397|AC002397 HMMgene1.1a STOP 11723 11725 0.295 - .emb|AC002397|AC002397 HMMgene1.1a ACC 10868 10869 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a DON 10813 10814 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 10191 10192 0.016 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 10191 10192 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a START 10135 10137 0.001 - .emb|AC002397|AC002397 HMMgene1.1a DON 10125 10126 0.007 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 10088 10089 0.552 - 0emb|AC002397|AC002397 HMMgene1.1a DON 10085 10086 0.006 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 10076 10078 0.002 - .emb|AC002397|AC002397 HMMgene1.1a DON 10043 10044 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 10042 10043 0.113 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 10036 10038 0.003 - .emb|AC002397|AC002397 HMMgene1.1a DON 10027 10028 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a DON 9964 9965 0.471 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 9960 9962 0.194 - .emb|AC002397|AC002397 HMMgene1.1a ACC 9650 9651 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 9646 9648 0.003 - .emb|AC002397|AC002397 HMMgene1.1a ACC 9268 9269 0.008 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 9249 9250 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 9194 9196 0.011 - .emb|AC002397|AC002397 HMMgene1.1a ACC 9133 9134 0.002 - 2emb|AC002397|AC002397 HMMgene1.1a STOP 9067 9069 0.002 - .emb|AC002397|AC002397 HMMgene1.1a ACC 8939 8940 0.005 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 8939 8940 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 8908 8910 0.002 - .emb|AC002397|AC002397 HMMgene1.1a STOP 8904 8906 0.005 - .emb|AC002397|AC002397 HMMgene1.1a ACC 8889 8890 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a DON 8835 8836 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 8637 8638 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a START 8630 8632 0.004 - .emb|AC002397|AC002397 HMMgene1.1a ACC 8622 8623 0.006 - 1emb|AC002397|AC002397 HMMgene1.1a DON 8514 8515 0.012 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 8415 8416 0.346 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 8325 8326 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a DON 8322 8323 0.004 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 8319 8321 0.001 - .emb|AC002397|AC002397 HMMgene1.1a STOP 8315 8317 0.342 - .emb|AC002397|AC002397 HMMgene1.1a ACC 8096 8097 0.019 - 1emb|AC002397|AC002397 HMMgene1.1a DON 8036 8037 0.006 - 1emb|AC002397|AC002397 HMMgene1.1a DON 7994 7995 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a DON 7989 7990 0.013 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 7868 7869 0.005 - 1emb|AC002397|AC002397 HMMgene1.1a DON 7790 7791 0.004 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 7600 7601 0.009 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 7503 7505 0.008 - .emb|AC002397|AC002397 HMMgene1.1a ACC 7071 7072 0.006 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 7071 7072 0.012 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 7024 7026 0.006 - .emb|AC002397|AC002397 HMMgene1.1a STOP 7001 7003 0.012 - .emb|AC002397|AC002397 HMMgene1.1a ACC 6958 6959 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 6864 6866 0.002 - .emb|AC002397|AC002397 HMMgene1.1a ACC 6681 6682 0.060 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 6622 6623 0.003 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 6521 6522 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 6491 6493 0.063 - .emb|AC002397|AC002397 HMMgene1.1a DON 6467 6468 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 6286 6287 0.004 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 6286 6287 0.001 - 2emb|AC002397|AC002397 HMMgene1.1a STOP 6274 6276 0.001 - .emb|AC002397|AC002397 HMMgene1.1a STOP 6269 6271 0.004 - .emb|AC002397|AC002397 HMMgene1.1a ACC 5771 5772 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 5712 5713 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a STOP 5638 5640 0.002 - .emb|AC002397|AC002397 HMMgene1.1a ACC 5514 5515 0.004 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 5483 5485 0.004 - .emb|AC002397|AC002397 HMMgene1.1a ACC 5133 5134 0.002 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 5093 5095 0.002 - .emb|AC002397|AC002397 HMMgene1.1a ACC 4917 4918 0.007 - 1emb|AC002397|AC002397 HMMgene1.1a ACC 4842 4843 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 4814 4816 0.009 - .emb|AC002397|AC002397 HMMgene1.1a ACC 4629 4630 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 4535 4537 0.003 - .emb|AC002397|AC002397 HMMgene1.1a ACC 3628 3629 0.002 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 3628 3629 0.005 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 3585 3587 0.005 - .emb|AC002397|AC002397 HMMgene1.1a STOP 3524 3526 0.002 - .emb|AC002397|AC002397 HMMgene1.1a ACC 1686 1687 0.001 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 1649 1651 0.002 - .emb|AC002397|AC002397 HMMgene1.1a ACC 127 128 0.001 - 0emb|AC002397|AC002397 HMMgene1.1a ACC 96 97 0.003 - 1emb|AC002397|AC002397 HMMgene1.1a STOP 77 79 0.005 - .
Genescan returns gene structure prediction, signals, and the peptides generates from the predicted gene structure.
GENSCAN 1.0 Date run: 29-Aug-102 Time: 06:06:14Sequence 06:06:12 : 20000 bp : 51.37% C+G : Isochore 3 (51 - 57 C+G%)Parameter matrix: HumanIso.smatPredicted genes/exons:Gn.Ex Type S .Begin ...End .Len Fr Ph I/Ac Do/T CodRg P.... Tscr..----- ---- - ------ ------ ---- -- -- ---- ---- ----- ----- ------ 1.01 Intr + 2651 2791 141 1 0 89 91 69 0.879 8.36 1.02 Intr + 3024 3164 141 2 0 86 9 174 0.189 10.36 1.03 Intr + 5458 5496 39 0 0 116 78 35 0.786 4.21 1.04 Intr + 6947 7053 107 1 2 89 50 89 0.816 4.71 1.05 Intr + 7133 7196 64 2 1 42 76 82 0.782 1.61 1.06 Intr + 7310 7472 163 1 1 72 99 113 0.930 10.86 1.07 Intr + 7578 7644 67 1 1 81 91 110 0.999 9.06 1.08 Intr + 7737 7938 202 0 1 100 56 150 0.980 12.81 1.09 Intr + 9071 9287 217 1 1 79 82 323 0.994 29.40 1.10 Term + 10080 10186 107 1 2 97 55 114 0.987 7.87 1.11 PlyA + 10420 10425 6 1.05 2.05 PlyA - 11605 11600 6 -1.95 2.04 Term - 11878 11723 156 1 0 117 49 152 0.974 12.45 2.03 Intr - 12297 12194 104 1 2 91 103 78 0.967 9.99 2.02 Intr - 12717 12424 294 1 0 78 92 198 0.820 16.73 2.01 Init - 13836 13776 61 0 1 44 41 5 0.027 -7.08 2.00 Prom - 13944 13905 40 -4.71 3.00 Prom + 14081 14120 40 -3.81 3.01 Init + 15468 15578 111 2 0 88 119 158 0.999 19.12 3.02 Intr + 18439 18564 126 0 0 92 82 145 0.999 15.68 3.03 Intr + 19685 19751 67 1 1 112 85 107 0.998 11.77 3.04 Intr + 19878 19977 100 1 1 122 -21 198 0.999 11.97
Predicted peptide sequence(s):Predicted coding sequence(s):>06:06:12|GENSCAN_predicted_peptide_1|415_aaAQVRPRCGRLVAFSSGGENPHGVWAVTRGRRCALALWHTWAPEHSEQEWTEAKELLQEEEEEEEEEDILSRDPSPEPPSHKLQRVQEKAGKPRRDARKACADITLAELVSGLEVVGRVQMRTRRTLRGHLAKIYAMHWATDSKLLVSASQDGKLIVWDTYTTNKVHAIPLRSSWVMTCAYAPSGNFVACGGLDNMCSIYNLKSREGNVKVSRELSAHTGYLSCCRFLDDNNIVTSSGDTTCALWDIETGQQKTVFVGHTGDCMSLAVSPDYKLFISGACDASAKLWDVREGTCRQTFTGHESDINAICFFPNGEAICTGSDDASCRLFDLRADQELTAYSQESIICGITSVAFSLSGRLLFAGYDDFNCNVWDSLKCERVGILSGHDNRVSCLGVTADGMAVATGSWDSFLKIWN>06:06:12|GENSCAN_predicted_CDS_1|1248_bpgctcaggtccgtcctcggtgtgggcgccttgtggccttcagctctggtggtgagaatccccatggtgtatgggctgtgactcggggacggcgctgtgccctagcactgtggcacacgtgggcacctgagcacagtgaacaggagtggacagaagccaaagagctgctgcaggaggaagaggaggaagaagaggaggaagacattctcagcagagacccttccccagaacccccaagtcacaagcttcagcgagtccaggagaaagctgggaagccccgccgggatgccaggaaagcctgtgcggacatcactctggctgagcttgtgtctggcctggaggtggtgggccgagtccagatgcggacacggaggacattaaggggacatctggccaagatctatgccatgcactgggccactgactctaagctgctcgtaagtgcctcgcaggatgggaagctgatcgtgtgggacacttataccaccaataaggtgcatgcaatcccgctgcgttcctcctgggtcatgacctgtgcctatgcaccatcagggaactttgtggcatgtggggggctggacaacatgtgttcaatctacaacctcaaatcccgcgagggcaatgtcaaggtcagccgggagctctctgctcacacaggttatctctcctgttgccgcttcctggatgacaacaacatcgtgactagctctggggacaccacatgtgccttgtgggacattgagacaggacagcagaagacagtgtttgtgggacacactggtgactgcatgagcctggctgtgtccccagactacaaactcttcatttcgggagcttgcgatgctagcgcgaagctctgggatgtgagggaagggacctgtcgtcagactttcactggccatgagtcagacatcaatgccatctgtttctttcccaacggggaggccatctgcactggctcagatgacgcctcctgccgcctctttgacctgagggcagaccaggaactgactgcctattcccaggagagcatcatctgcggcatcacttcagtagccttctcgctcagtgggcgcctgctctttgcaggctatgatgacttcaactgcaatgtctgggactctctgaagtgcgagcgtgtaggcatactctctggccatgacaacagagtcagctgcctgggggtcactgctgacggcatggctgtggccactggttcctgggacagcttcctcaaaatctggaactga>06:06:12|GENSCAN_predicted_peptide_2|204_aaMCHYRAWQLSAFFNLHWAVADPQCSLVKELSEVLETEASESISSPELALPRETPLFYDLDLSSDPQLSPEDQLLPWSQAELDPKQVFTKEEAKQSAETIAASQNSDKPSRDPETPQSSGSKRSRRKANSKVLGRSPLTILQDDNSPGTLTLRQGKRPSALSENVKDLKEGVVLGTGRFLKAGGGAREPNQDHDKENQHFALLES>06:06:12|GENSCAN_predicted_CDS_2|615_bpatgtgccattaccgtgcctggcaactttctgcattctttaacctgcactgggcagtcgcagaccctcagtgctcactggtgaaagagctgagcgaagtattggagacagaagcgtcggaatcgatttcctccccagagcttgctctgccccgggaaacgcctttattttatgacctggacctgtcttcagatcctcagttatcccctgaggaccagttactgccttggagccaggctgaactcgatcccaaacaggtgtttaccaaggaggaagccaaacaatccgcagaaactatagctgccagccagaactcagacaagccctccagagacccagagactccccagtcctcaggttctaagcgcagcagacgaaaagcaaacagcaaggttctagggaggtcccctctcaccatcctgcaggatgacaactcccctgggaccttgacactacgacagggtaagcggccttctgccctcagtgagaacgttaaggacctaaaggaaggagtcgttcttggaactggaagatttctcaaagctggaggaggagcacgggagccaaaccaggaccacgacaaggaaaatcagcattttgccttgttggagagctag>06:06:12|GENSCAN_predicted_peptide_3|135_aaMAELSEEALLSVLPTIRVPKAGDRVHKDECAFSFDTPESEGGLYICMNTFLGFGKQYVERHFNKTGQRVYLHLRRTRRPKEEDTSAGTGDPPRKKPTRLAIGVEGGFDLTEDKFEFDEDVKIVILPDYLEIARDX>06:06:12|GENSCAN_predicted_CDS_3|405_bpatggcggagctgagtgaagaggcgctgctgtcagtgttaccgacgatccgtgtccccaaggcgggagaccgggtccataaagacgagtgcgctttctctttcgacacgccggagtctgagggtggcctctatatctgcatgaacacattcctgggattcgggaagcagtatgtggagagacacttcaacaagacaggccagcgtgtctacctgcacctccggaggacccggcgaccgaaagaagaggacaccagtgcaggcactggagacccacctcggaagaagcccacccggctggccattggtgttgaaggagggtttgacctcaccgaggacaagtttgaatttgacgaggatgtgaagattgtcattttgcccgattacctggagatcgctcgggatggnExplanationGn.Ex : gene number, exon number (for reference)Type : Init = Initial exon (ATG to 5' splice site) Intr = Internal exon (3' splice site to 5' splice site) Term = Terminal exon (3' splice site to stop codon) Sngl = Single-exon gene (ATG to stop) Prom = Promoter (TATA box / initation site) PlyA = poly-A signal (consensus: AATAAA)S : DNA strand (+ = input strand; - = opposite strand)Begin : beginning of exon or signal (numbered on input strand)End : end point of exon or signal (numbered on input strand)Len : length of exon or signal (bp)Fr : reading frame (a forward strand codon ending at x has frame x mod 3)Ph : net phase of exon (exon length modulo 3)I/Ac : initiation signal or 3' splice site score (tenth bit units)Do/T : 5' splice site or termination signal score (tenth bit units)CodRg : coding region score (tenth bit units)P : probability of exon (sum over all parses containing exon)Tscr : exon score (depends on length, I/Ac, Do/T and CodRg scores)CommentsThe SCORE of a predicted feature (e.g., exon or splice site) is alog-odds measure of the quality of the feature based on local sequenceproperties. For example, a predicted 5' splice site withscore > 100 is strong; 50-100 is moderate; 0-50 is weak; andbelow 0 is poor (more than likely not a real donor site).The PROBABILITY of a predicted exon is the estimated probability underGENSCAN's model of genomic sequence structure that the exon is correct.This probability depends in general on global as well as local sequenceproperties, e.g., it depends on how well the exon fits with neighboringexons. It has been shown that predicted exons with higher probabilitiesare more likely to be correct than those with lower probabilities.
Genie allow to run multiple searches in one go (multiple FASTA sequences).
The results are returned by email. They are very concise (scores are attributed to the full predicton, but not for each single exon).
Genie's gene predictions for sequence unknown:Forward strand (score = 1025.8):Exon 0: 1545- 1586Exon 1: 2651- 2791Exon 2: 5458- 5496Exon 3: 7310- 7472Exon 4: 7578- 7644Exon 5: 7737- 7938Exon 6: 9071- 9287Exon 7: 10080-10186Reverse strand (score = 944.6):Exon 0: 14755-14590Exon 1: 14499-14446Exon 2: 12642-12424Exon 3: 12297-12194Exon 4: 4917- 4788Exon 5: 2774- 2653---------------------Note: the higher (more positive) the score, the more likely there isto be a real gene on that strand.References:1. D. Kulp, D. Haussler, M.G. Reese, and F.H. Eeckman (1996).A generalized Hidden Markov Model for the recognition of human genes in DNA.ISMB-96, St. Louis, MO, AAAI/MIT Press.2. M.G. Reese, F.H. Eeckman, D. Kulp, D. Haussler (1997).Improved splice site detection in Genie.Proceedings of the First Annual International Conference on ComputationalMolecular Biology (RECOMB), 1997, Santa Fe, NM, ACM Press, New York.
You can het more information about Geneid from here.
The following output has been produced using the standard options.
## gff-version 2## date Thu Aug 29 13:25:45 2002## source-version: geneid v 1.1 -- geneid@imim.es## Sequence unknown - Length = 20000 bps# Optimal Gene Structure. 3 genes. Score = 43.130057 # Gene 1 (Forward). 8 exons. 393 aa. Score = 19.805313 unknown geneid_v1.1 Internal 2557 2791 1.92 + 1 1unknown geneid_v1.1 Internal 3024 3170 2.13 + 0 1unknown geneid_v1.1 Internal 5458 5496 2.80 + 0 1unknown geneid_v1.1 Internal 7310 7472 2.35 + 0 1unknown geneid_v1.1 Internal 7578 7644 1.78 + 2 1unknown geneid_v1.1 Internal 7737 7938 0.99 + 1 1unknown geneid_v1.1 Internal 9071 9287 7.25 + 0 1unknown geneid_v1.1 Terminal 10080 10186 0.59 + 2 1# Gene 2 (Reverse). 3 exons. 282 aa. Score = 7.818153 unknown geneid_v1.1 Terminal 11723 11878 1.09 - 0 2unknown geneid_v1.1 Internal 12194 12717 8.21 - 2 2unknown geneid_v1.1 First 14590 14755 -1.48 - 0 2# Gene 3 (Forward). 4 exons. 134 aa. Score = 15.506592 unknown geneid_v1.1 First 15468 15578 5.82 + 0 3unknown geneid_v1.1 Internal 18439 18564 3.64 + 0 3unknown geneid_v1.1 Internal 19685 19751 2.47 + 0 3unknown geneid_v1.1 Internal 19878 19977 3.59 + 2 3
GeneBuilder is the only service presented here than can use homolgy information (proteins, ESTs, CDSs) to help the prediction of the coding regions.
This information can be added, for example, after a first ab inition prediction and homology search.The output obtained from GeneBuilder using the standard options and no homology information.
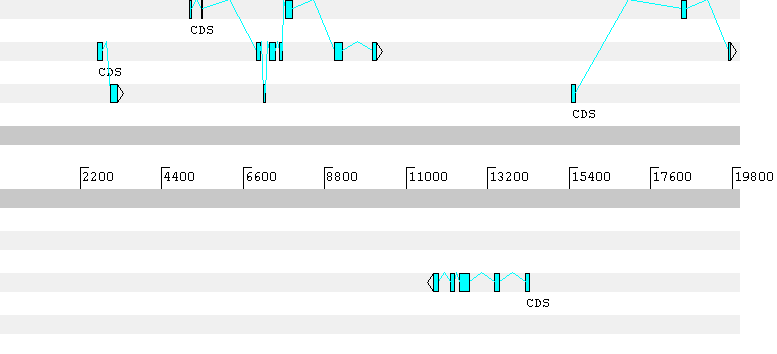
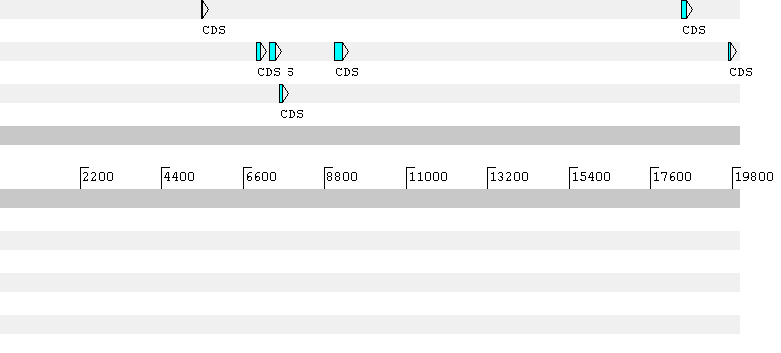
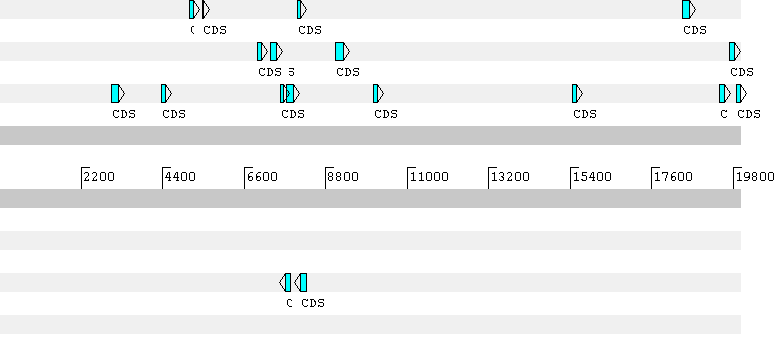
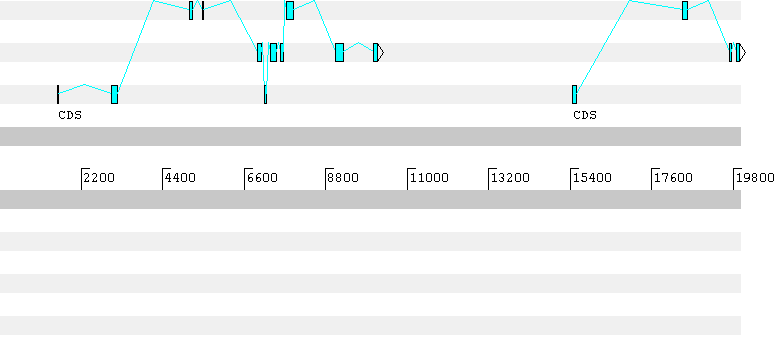
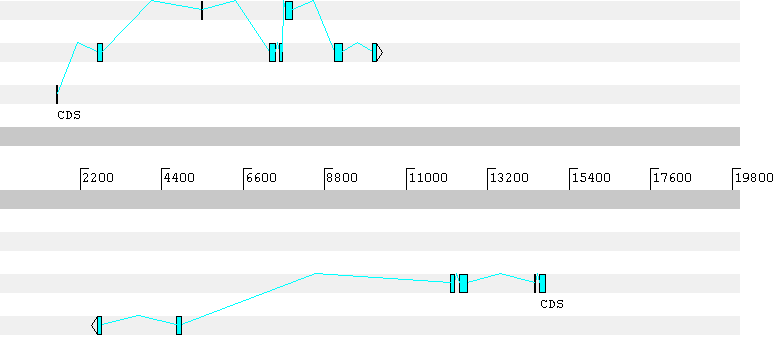
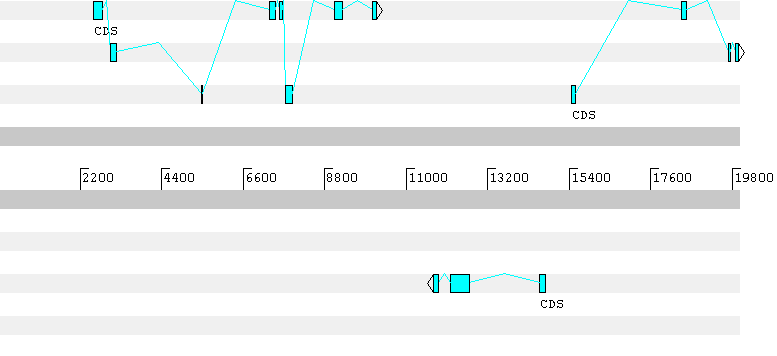
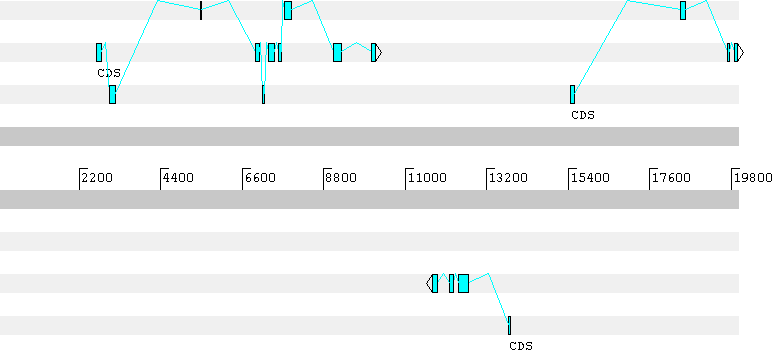
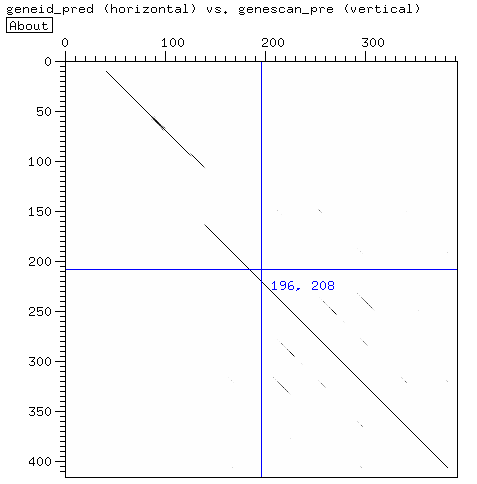
The first gene predicted by Geneid has a frameshift in the prediction. Here the predicted gene sequence:
>unknownGGCCAGTTTCTCATTTGCCCTAAACTCGTCCCTGAGTGAGGGAGGGCAGAGTAAGAGAATCAGGAAGCCTGATGCTGTGTTCCTGCATTCTCAGGCTCAGGTCCGTCCTCGGTGTGGGCGCCTTGTGGCCTTCAGCTCTGGTGGTGAGAATCCCCATGGTGTATGGGCTGTGACTCGGGGACGGCGCTGTGCCCTAGCACTGTGGCACACGTGGGCACCTGAGCACAGTGAACAGGAGTGGACAGAAGCCAAAGAGCTGCTGCAGGAGGAAGAGGAGGAAGAAGAGGAGGAAGACATTCTCAGCAGAGACCCTTCCCCAGAACCCCCAAGTCACAAGCTTCAGCGAGTCCAGGAGAAAGCTGGGAAGCCCCGCCGGGTCCGGGATGCCAGGAAAGCCTGTGCGGACATCACTCTGGCTGAGGTGCATGCAATCCCGCTGCGTTCCTCCTGGGTCATGACCTGTGCCTATGCACCATCAGGGAACTTTGTGGCATGTGGGGGGCTGGACAACATGTGTTCAATCTACAACCTCAAATCCCGCGAGGGCAATGTCAAGGTCAGCCGGGAGCTCTCTGCTCACACAGGTTATCTCTCCTGTTGCCGCTTCCTGGATGACAACAACATCGTGACTAGCTCTGGGGACACCACATGTGCCTTGTGGGACATTGAGACAGGACAGCAGAAGACAGTGTTTGTGGGACACACTGGTGACTGCATGAGCCTGGCTGTGTCCCCAGACTACAAACTCTTCATTTCGGGAGCTTGCGATGCTAGCGCGAAGCTCTGGGATGTGAGGGAAGGGACCTGTCGTCAGACTTTCACTGGCCATGAGTCAGACATCAATGCCATCTGTTTCTTTCCCAACGGGGAGGCCATCTGCACTGGCTCAGATGACGCCTCCTGCCGCCTCTTTGACCTGAGGGCAGACCAGGAACTGACTGCCTATTCCCAGGAGAGCATCATCTGCGGCATCACTTCAGTAGCCTTCTCGCTCAGTGGGCGCCTGCTCTTTGCAGGCTATGATGACTTCAACTGCAATGTCTGGGACTCTCTGAAGTGCGAGCGTGTAGGCATACTCTCTGGCCATGACAACAGAGTCAGCTGCCTGGGGGTCACTGCTGACGGCATGGCTGTGGCCACTGGTTCCTGGGACAGCTTCCTCAAAATCTGGAACTGA
... and its protein.
ASFSFALNSSLSEGGQSKRIRKPDAVFLHSQAQVRPRCGRLVAFSSGGENPHGVWAVTRGRRCALALWHTWAPEHSEQEWTEAKELLQEEEEEEEEEDILSRDPSPEPPSHKLQRVQEKAGKPRRVRDARKACADITLAEVHAIPLRSSWVMTCAYAPSGNFVACGGLDNMCSIYNLKSREGNVKVSRELSAHTGYLSCCRFLDDNNIVTSSGDTTCALWDIETGQQKTVFVGHTGDCMSLAVSPDYKLFISGACDASAKLWDVREGTCRQTFTGHESDINAICFFPNGEAICTGSDDASCRLFDLRADQELTAYSQESIICGITSVAFSLSGRLLFAGYDDFNCNVWDSLKCERVGILSGHDNRVSCLGVTADGMAVATGSWDSFLKIWN
This is the Genescan protein prediction.
AQVRPRCGRLVAFSSGGENPHGVWAVTRGRRCALALWHTWAPEHSEQEWTEAKELLQEEEEEEEEEDILSRDPSPEPPSHKLQRVQEKAGKPRRDARKACADITLAELVSGLEVVGRVQMRTRRTLRGHLAKIYAMHWATDSKLLVSASQDGKLIVWDTYTTNKVHAIPLRSSWVMTCAYAPSGNFVACGGLDNMCSIYNLKSREGNVKVSRELSAHTGYLSCCRFLDDNNIVTSSGDTTCALWDIETGQQKTVFVGHTGDCMSLAVSPDYKLFISGACDASAKLWDVREGTCRQTFTGHESDINAICFFPNGEAICTGSDDASCRLFDLRADQELTAYSQESIICGITSVAFSLSGRLLFAGYDDFNCNVWDSLKCERVGILSGHDNRVSCLGVTADGMAVATGSWDSFLKIWN
Althoug there is a frameshift in the gene predicted by Geneid, the protein is correcly translated from the 2nd frame. Using Dotlet or LALIGN, you can easily obseve the difference between the 2 prediction, especially the missing exon from the Geneid prediction.