The grid should be 4 times 7x7 spots.
From ScanAlyze manual: Recommended Procedure for Gridding Arrays
1) Load Images (red and green channels)
2) Adjust Gain so the most spots are visible against the background
you can save the image
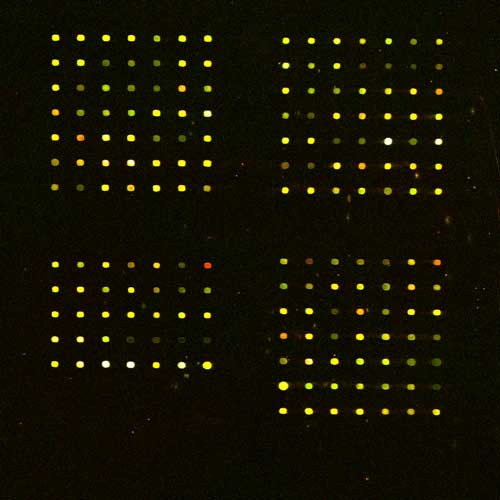
3) Create all grids at once with values obtained from array manufacture
distance between grids 5000, distance between spots 500, spot size 18
4) Hide grids
5) Move and stretch each grid box so that the corners are close to their appropriate location on the image
6) Show grids
7) Save grids
8) Refine (either by selecting one grid at a time or by selecting all grids) until positions look correct
9) Save grids
10) Move individual spots whose positions need correction
11) Save grids
12) Flag spots in problem areas
13) Save grids
14) Calculate and save data (see below)
Steps 1-9 take an experienced user approximately 2 minutes on a reasonably fast computer. Flagging can be a time consuming process. Methods are being tested to automatically locate poor quality spots and will be included in future versions. Many quality control parameters are provided in the current version. By using these values, users may find most flagging unnecessary.
2) export the data and open Excel to visualize the numbers... Then use the files "table.xls" and "gene-list_sorted.xls" to help yourself to identify the genes.
3) try to apply a very simple normalisation using a global Red vs Green normalisation.
4) try the Def664 and Def665 images. What happend? In 664 the covering glas was misplaced, the hybridization went bad. In 665 the covering glas was broken...