Multiple Alignments, Patterns and PSI-BLAST
1) Blast the full sequence of Y33K_HUMAN against Swissprot
BLASTP 2.2.1 [Jul-12-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= sp|Q04323|Y33K_HUMAN Hypothetical 33.4 kDa protein
(298 letters)
Database: swiss
102,164 sequences; 37,554,368 total letters
Searching..................................................done
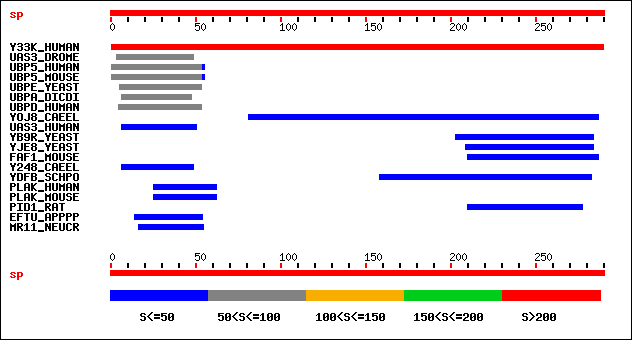 Score E
Sequences producing significant alignments: (bits) Value
sp|Q04323|Y33K_HUMAN Hypothetical 33.4 kDa protein.[Homo sapiens] 387 e-108
sp|Q9VCE9|UAS3_DROME (CG13604)UBASH3A protein homolog.[Drosophil... 64 3e-10
sp|P56399|UBP5_MOUSE (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P45974|UBP5_HUMAN (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P38237|UBPE_YEAST (UBP14..)Ubiquitin carboxyl-terminal hydrol... 57 5e-08
sp|Q92995|UBPD_HUMAN (USP13..)Ubiquitin carboxyl-terminal hydrol... 51 2e-06
sp|P54201|UBPA_DICDI (UBPA)Ubiquitin carboxyl-terminal hydrolase... 51 2e-06
sp|P34631|YOJ8_CAEEL (ZK353.8)Hypothetical 51.6 kDa protein ZK35... 50 3e-06
sp|P57075|UAS3_HUMAN (UBASH3A)UBASH3A protein.[Homo sapiens] 49 7e-06
sp|P38349|YB9R_YEAST (YBR273C..)Hypothetical 50.0 kDa protein in... 47 3e-05
sp|P47049|YJE8_YEAST (YJL048C..)Hypothetical 45.0 kDa protein in... 40 0.005
sp|P54731|FAF1_MOUSE (FAF1)FAF1 protein (FAS-associated factor 1... 38 0.022
sp|O76387|Y248_CAEEL (C24G6.8)Hypothetical 33.7 kDa protein C24G... 36 0.065
sp|Q10483|YDFB_SCHPO (SPAC17C9.11C)Hypothetical 27.6 kDa protein... 31 2.1
sp|P10688|PID1_RAT (PLCD1)1-phosphatidylinositol-4,5-bisphosphat... 30 3.6
sp|Q02257|PLAK_MOUSE (JUP)Junction plakoglobin (Desmoplakin III)... 30 4.7
sp|P14923|PLAK_HUMAN (JUP..)Junction plakoglobin (Desmoplakin II... 30 4.7
sp|Q9ZEU3|EFTU_APPPP (TUF)Elongation factor Tu (EF-Tu).[Apple pr... 30 4.7
sp|Q9C291|MR11_NEUCR (MUS-23..)Double-strand break repair protei... 30 6.1
Score E
Sequences producing significant alignments: (bits) Value
sp|Q04323|Y33K_HUMAN Hypothetical 33.4 kDa protein.[Homo sapiens] 387 e-108
sp|Q9VCE9|UAS3_DROME (CG13604)UBASH3A protein homolog.[Drosophil... 64 3e-10
sp|P56399|UBP5_MOUSE (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P45974|UBP5_HUMAN (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P38237|UBPE_YEAST (UBP14..)Ubiquitin carboxyl-terminal hydrol... 57 5e-08
sp|Q92995|UBPD_HUMAN (USP13..)Ubiquitin carboxyl-terminal hydrol... 51 2e-06
sp|P54201|UBPA_DICDI (UBPA)Ubiquitin carboxyl-terminal hydrolase... 51 2e-06
sp|P34631|YOJ8_CAEEL (ZK353.8)Hypothetical 51.6 kDa protein ZK35... 50 3e-06
sp|P57075|UAS3_HUMAN (UBASH3A)UBASH3A protein.[Homo sapiens] 49 7e-06
sp|P38349|YB9R_YEAST (YBR273C..)Hypothetical 50.0 kDa protein in... 47 3e-05
sp|P47049|YJE8_YEAST (YJL048C..)Hypothetical 45.0 kDa protein in... 40 0.005
sp|P54731|FAF1_MOUSE (FAF1)FAF1 protein (FAS-associated factor 1... 38 0.022
sp|O76387|Y248_CAEEL (C24G6.8)Hypothetical 33.7 kDa protein C24G... 36 0.065
sp|Q10483|YDFB_SCHPO (SPAC17C9.11C)Hypothetical 27.6 kDa protein... 31 2.1
sp|P10688|PID1_RAT (PLCD1)1-phosphatidylinositol-4,5-bisphosphat... 30 3.6
sp|Q02257|PLAK_MOUSE (JUP)Junction plakoglobin (Desmoplakin III)... 30 4.7
sp|P14923|PLAK_HUMAN (JUP..)Junction plakoglobin (Desmoplakin II... 30 4.7
sp|Q9ZEU3|EFTU_APPPP (TUF)Elongation factor Tu (EF-Tu).[Apple pr... 30 4.7
sp|Q9C291|MR11_NEUCR (MUS-23..)Double-strand break repair protei... 30 6.1
- What are the positive matches? They are shown in green
- Do you detect a potential domain? Yes in the first 60 residues of the query
- What happens if you remove the filter (uncheck BLAST filter button)? The results are polluted by parasite sequences matching in a glutamic acid rich region
2) Select the matching sequences and create a multiple fasta format file by cut&paste
>y33k_human
MAELTALESLIEMGFPRGRAEKALALTGNQGIEAAMDWLMEHEDDPDVDEPLETPLGHIL
>uas3_drome
LTPLQTLLQMGFPRHRAEKALASTGNRGVQIASDWLLAHVNDGTLDE
>ubp5_mouse1
MLDESVIIQLVEMGFPMDACRKAVYYTGNSGAEAAMNWVMSHMDDPDFANPLILP
>ubp5_mouse2
TIVSMGFSRDQALKALRATNNSLERAVDWIFSHIDDLDAEAAMDISEG
>ubp5_human1
MLDESVIIQLVEMGFPMDACRKAVYYTGNSGAEAAMNWVMSHMDDPDFANPLILP
>ubp5_human2
TIVSMGFSRDQALKALRATNNSLERAVDWIFSHIDDLDAEAAMDISEG
>ubpe_yeast
SISQLIEMGFTQNASVRALFNTGNQDAESAMNWLFQHMDDPDLNDPFVPP
>ubpd_human1
SSVMQLAEMGFPLEACRKAVYFTGNMGAEVAFNWIIVHMEEPDFAEPLTMP
>ubpd_human2
ITSMGFQRNQAIQALRATNNN-LERALDWIFSHPEFEEDSD
>ubpa_dicdi1
LDTLLSMDFPLVRCKKALLATGGKDAELAMNWIFEHTEDPDID
>ubpa_dicdi2
VDNIIGMGFTDSQAKLALKNTKGNLERAADWLFSHIDNLD
>uas3_human
LEPLLAMGFPVHTALKALAATGRKTAEEALAWLHDHCNDPSLDDPI
then perform a multiple alignment using ClustalW (or Emma on command-line)
CLUSTAL W (1.74) multiple sequence alignment
y33k_human MAELTALESLIEMGFPRGRAEKALALTGNQGIEAAMDWLMEHEDDPDVDEPLETPLGHIL
uas3_drome ---LTPLQTLLQMGFPRHRAEKALASTGNRGVQIASDWLLAHVNDGTLDE----------
ubp5_mouse1 MLDESVIIQLVEMGFPMDACRKAVYYTGNSGAEAAMNWVMSHMDDPDFANPLILP-----
ubp5_mouse2 --------TIVSMGFSRDQALKALRATNNS-LERAVDWIFSHIDDLDAEAAMDISEG---
ubp5_human1 MLDESVIIQLVEMGFPMDACRKAVYYTGNSGAEAAMNWVMSHMDDPDFANPLILP-----
ubp5_human2 --------TIVSMGFSRDQALKALRATNNS-LERAVDWIFSHIDDLDAEAAMDISEG---
ubpe_yeast -----SISQLIEMGFTQNASVRALFNTGNQDAESAMNWLFQHMDDPDLNDPFVPP-----
ubpd_human1 ----SSVMQLAEMGFPLEACRKAVYFTGNMGAEVAFNWIIVHMEEPDFAEPLTMP-----
ubpd_human2 ---------ITSMGFQRNQAIQALRATNNN-LERALDWIFSHP---EFEEDSD-------
ubpa_dicdi1 ------LDTLLSMDFPLVRCKKALLATGGKDAELAMNWIFEHTEDPDID-----------
ubpa_dicdi2 ------VDNIIGMGFTDSQAKLALKNTKGN-LERAADWLFSHIDNLD-------------
uas3_human ------LEPLLAMGFPVHTALKALAATGRKTAEEALAWLHDHCNDPSLDDPI--------
: *.* . *: * : * *: *
- How many conserved residues do you see (count the stars)? 7 conserved residues
3) Create a simple PROSITE pattern and search (or here or fuzzpro on command-line) against Swissprot
M-[GD]-F-x(4)-[SAC]-x(2)-A-[LV]-x(2)-T-x(4,5)-[EQ]-x-A-x(2)-W-[LIV]-x(2)-H
- Do you detect more proteins? No
- Why? Because of the stringence of the pattern, one should start with a relaxed pattern and increase the stringency little by little to remove false positives
4) Do a PSI-BLAST (or here) against SwissProt with the first 60 residues of Y33K_HUMAN
- Iterate 3-4 times by selecting potential matches
- Do you detect more proteins? Yes
- Why? Because the PSI-BLAST is more flexible in allowing mismatches and amino acid substitutions
- What is the conserved domain? It is a Ubiquitin Associated domain (UBA) found in many proteins involved in the ubiquitin pathway. It is known to bind ubiquitin.
 Score E
Sequences producing significant alignments: (bits) Value
sp|Q04323|Y33K_HUMAN Hypothetical 33.4 kDa protein.[Homo sapiens] 387 e-108
sp|Q9VCE9|UAS3_DROME (CG13604)UBASH3A protein homolog.[Drosophil... 64 3e-10
sp|P56399|UBP5_MOUSE (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P45974|UBP5_HUMAN (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P38237|UBPE_YEAST (UBP14..)Ubiquitin carboxyl-terminal hydrol... 57 5e-08
sp|Q92995|UBPD_HUMAN (USP13..)Ubiquitin carboxyl-terminal hydrol... 51 2e-06
sp|P54201|UBPA_DICDI (UBPA)Ubiquitin carboxyl-terminal hydrolase... 51 2e-06
sp|P34631|YOJ8_CAEEL (ZK353.8)Hypothetical 51.6 kDa protein ZK35... 50 3e-06
sp|P57075|UAS3_HUMAN (UBASH3A)UBASH3A protein.[Homo sapiens] 49 7e-06
sp|P38349|YB9R_YEAST (YBR273C..)Hypothetical 50.0 kDa protein in... 47 3e-05
sp|P47049|YJE8_YEAST (YJL048C..)Hypothetical 45.0 kDa protein in... 40 0.005
sp|P54731|FAF1_MOUSE (FAF1)FAF1 protein (FAS-associated factor 1... 38 0.022
sp|O76387|Y248_CAEEL (C24G6.8)Hypothetical 33.7 kDa protein C24G... 36 0.065
sp|Q10483|YDFB_SCHPO (SPAC17C9.11C)Hypothetical 27.6 kDa protein... 31 2.1
sp|P10688|PID1_RAT (PLCD1)1-phosphatidylinositol-4,5-bisphosphat... 30 3.6
sp|Q02257|PLAK_MOUSE (JUP)Junction plakoglobin (Desmoplakin III)... 30 4.7
sp|P14923|PLAK_HUMAN (JUP..)Junction plakoglobin (Desmoplakin II... 30 4.7
sp|Q9ZEU3|EFTU_APPPP (TUF)Elongation factor Tu (EF-Tu).[Apple pr... 30 4.7
sp|Q9C291|MR11_NEUCR (MUS-23..)Double-strand break repair protei... 30 6.1
Score E
Sequences producing significant alignments: (bits) Value
sp|Q04323|Y33K_HUMAN Hypothetical 33.4 kDa protein.[Homo sapiens] 387 e-108
sp|Q9VCE9|UAS3_DROME (CG13604)UBASH3A protein homolog.[Drosophil... 64 3e-10
sp|P56399|UBP5_MOUSE (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P45974|UBP5_HUMAN (USP5..)Ubiquitin carboxyl-terminal hydrola... 60 6e-09
sp|P38237|UBPE_YEAST (UBP14..)Ubiquitin carboxyl-terminal hydrol... 57 5e-08
sp|Q92995|UBPD_HUMAN (USP13..)Ubiquitin carboxyl-terminal hydrol... 51 2e-06
sp|P54201|UBPA_DICDI (UBPA)Ubiquitin carboxyl-terminal hydrolase... 51 2e-06
sp|P34631|YOJ8_CAEEL (ZK353.8)Hypothetical 51.6 kDa protein ZK35... 50 3e-06
sp|P57075|UAS3_HUMAN (UBASH3A)UBASH3A protein.[Homo sapiens] 49 7e-06
sp|P38349|YB9R_YEAST (YBR273C..)Hypothetical 50.0 kDa protein in... 47 3e-05
sp|P47049|YJE8_YEAST (YJL048C..)Hypothetical 45.0 kDa protein in... 40 0.005
sp|P54731|FAF1_MOUSE (FAF1)FAF1 protein (FAS-associated factor 1... 38 0.022
sp|O76387|Y248_CAEEL (C24G6.8)Hypothetical 33.7 kDa protein C24G... 36 0.065
sp|Q10483|YDFB_SCHPO (SPAC17C9.11C)Hypothetical 27.6 kDa protein... 31 2.1
sp|P10688|PID1_RAT (PLCD1)1-phosphatidylinositol-4,5-bisphosphat... 30 3.6
sp|Q02257|PLAK_MOUSE (JUP)Junction plakoglobin (Desmoplakin III)... 30 4.7
sp|P14923|PLAK_HUMAN (JUP..)Junction plakoglobin (Desmoplakin II... 30 4.7
sp|Q9ZEU3|EFTU_APPPP (TUF)Elongation factor Tu (EF-Tu).[Apple pr... 30 4.7
sp|Q9C291|MR11_NEUCR (MUS-23..)Double-strand break repair protei... 30 6.1